Comparative Karyotype Analysis and Chromosome Evolution in the Genus Ocimum L. from Thailand
DOI:
https://doi.org/10.58837/tnh.21.1.247165Keywords:
basil, chromosomal rearrangement, diploidization, genome downsizing, karyotype asymmetry, polyploidyAbstract
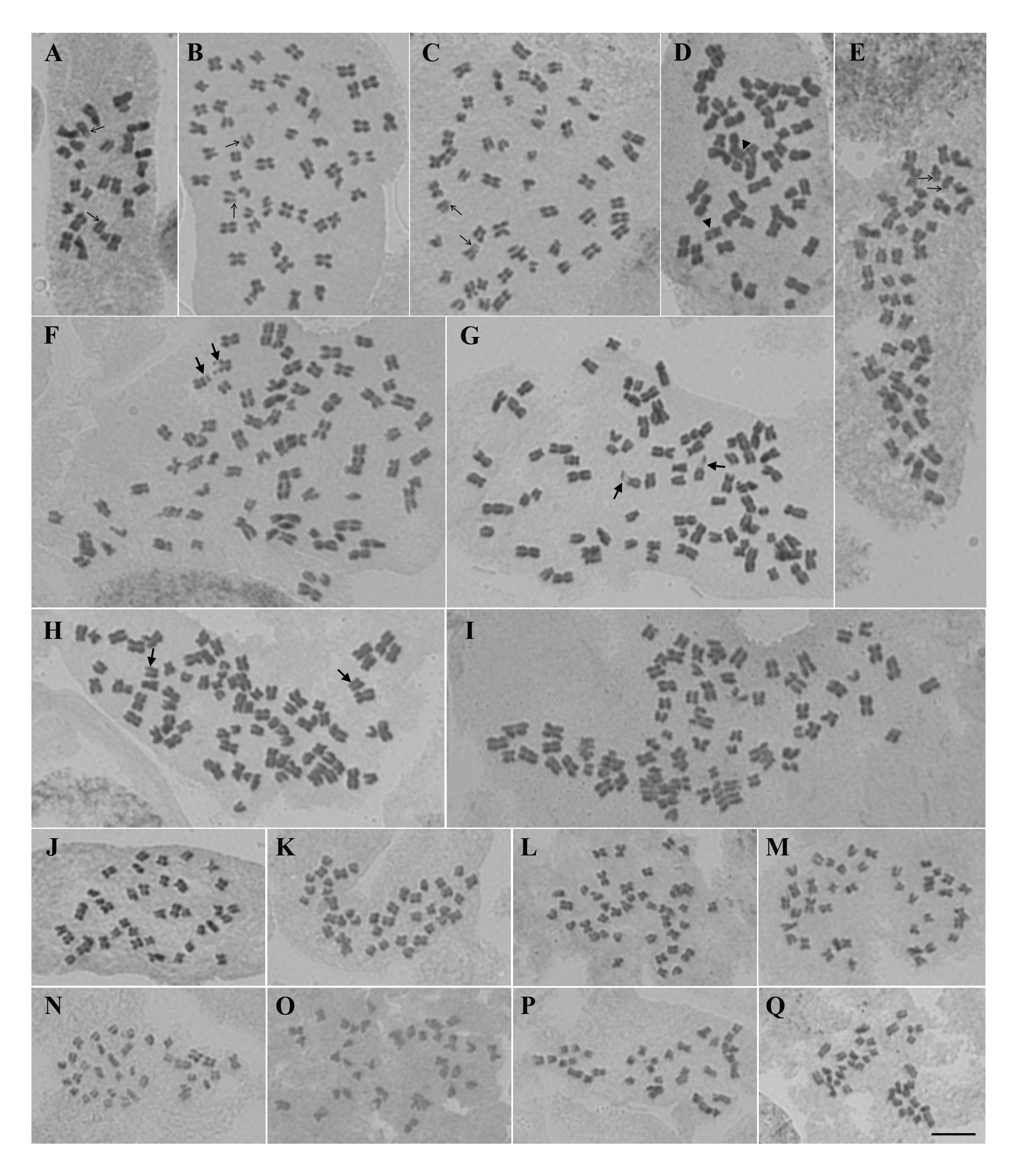
Ocimum L. (Lamiaceae) is one of the best-known genera of aromatic herbs in the world for its economic and medicinal importance. Most species are polyploids, harboring a large variation in genome and chromosome compositions. This study reports the comparative karyotype analysis among O. americanum L. and four accessions each of O. basilicum L., O. africanum Lour., O. gratissimum L. and O. tenuiflorum L. from Thailand. We found chromosome numbers of O. americanum, O. basilicum, O. africanum, O. gratissimum and O. tenuiflorum to be 2n=26, 52, 78, 40 and 36, respectively. All these species have asymmetric karyotypes, but O. tenuiflorum exhibited higher level of asymmetry than the other species. Although the chromosome numbers of all the studied species were stable among accessions, differences were found in the karyotypic constitutions. Intra-specific variation in karyotype formulae and satellite chromosomes of O. basilicum and O. africanum suggest that chromosomal rearrangements are involved in the chromosome evolution of these karyotypically unstable species, indicating that they may be newly formed polyploids. On the other hand, the disproportionate rise between ploidy level and the total karyotype length in the derived and stable polyploids, O. gratissimum and O. tenuiflorum, points towards a process of diploidization via genome downsizing.
References
Alix, K., Gérard, P.R., Schwarzacher, T. and Heslop-Harrison, J.S.P. 2017. Polyploidy and interspecific hybridization: Partners for adaptation, speciation and evolution in plants. Annals of Botany, 120: 183-194.
Avetisyan, A., Markosian, A., Petrosyan, M., Sahakyan, N., Babayan, A., Aloyan, S. and Trchounian, A. 2017. Chemical composition and some biological activities of the essential oils from basil Ocimum different cultivars. BMC Complementary and Alternative Medicine, 17: 60-67.
Battaglia, E. 1955. Chromosome morphology and terminology. Caryologia, 8: 179-187.
Berić, T., Nikolić, B., Stanojević, J., Vuković-Gačić, B. and Knežević-Vukčević, J. 2008. Protective effect of basil (Ocimum basilicum L.) against oxidative DNA damage and mutagenesis. Food and Chemical Toxicology, 46: 724-732.
Bir, S.S. and Saggoo, M.I.S. 1985. Cytological studies on members of family Labiatae from Kodaikanal and adjoining areas (South India). Proceedings: Plant Sciences, 94: 619-626.
Bomblies, K., Jones, G., Franklin, C., Zickler, D. and Kleckner, N. 2016. The challenge of evolving stable polyploidy: Could an increase in "crossover interference distance" play a central role?. Chromosoma, 125: 287-300.
Carović-Stanko, K., Liber, Z., Besendorfer, V., Javornik, B., Bohanec, B., Kolak, I. and Satovic, Z. 2010. Genetic relations among basil taxa (Ocimum L.) based on molecular markers, nuclear DNA content, and chromosome number. Plant Systematics and Evolution, 285: 13-22.
Christina, V.L.P. and Annamalai, A. 2014. Nucleotide based validation of Ocimum species by evaluating three candidate barcodes of the chloroplast region. Molecular Ecology Resources, 14: 60-68.
Dash, C., Afroz, M., Sultana, S. and Alam, S. 2017. Conventional and fluorescent karyotype analysis of Ocimum spp. Cytologia, 82: 429-434.
Dhasmana, A. 2013. Somatic chromosomal studies in Ocimum basilicum & Ocimum sanctum L. International Journal of Phytomedicine, 5: 330-340.
Edet, O.U. and Aikpokpodion, P.O. 2014. Karyotype analysis of Ocimum basilicum in South-Eastern Nigeria. American Journal of Plant Sciences, 5: 126-131.
Garcia, S., Kovařík, A., Leitch, A.R. and Garnatje, T. 2017. Cytogenetic features of rRNA genes across land plants: Analysis of the plant rDNA database. The Plant Journal, 89: 1020-1030.
Hoffmann, A. and Rieseberg, L. 2008. Revisiting the impact of inversions in evolution: From population genetic markers to drivers of adaptive shifts and speciation?. Annual Review of Ecology, Evolution, and Systematics, 39: 21-42.
Hussain, A.I., Anwar, F., Hussain Sherazi, S.T. and Przybylski, R. 2008. Chemical composition, antioxidant and antimicrobial activities of basil (Ocimum basilicum) essential oils depends on seasonal variations. Food Chemistry, 108: 986-995.
Idowu, J. and Oziegbe, M. 2017. Mitotic and meiotic studies on two species of Ocimum (Lamiaceae) and their F1 hybrids. Botanica Lithuanica, 23: 59-67.
Khosla, M.K. 1988. Cytomorphological study of the F2 variants of F1 hybrids of Ocimum gratissimum L. (2n=40) and Ocimum viride Willd. (2n=40). Cytologia, 53: 561-570.
Khosla, M.K. 1989. Chromosome meiotic behaviour and ploidy nature in genus Ocimum (Lamiaceae), "Sanctum Group". Cytologia, 54: 223-229.
Khosla, M.K. 1995. Study of inter-relationship, phylogeny and evolutionary tendencies in genus Ocimum. Indian Journal of Genetics and Plant Breeding, 55: 71-83.
Kumar, A., Mishra, P., Baskaran, K., Shukla, A.K., Shasany, A.K. and Sundaresan, V. 2016. Higher efficiency of ISSR markers over plastid psbA-trnH region in resolving taxonomical status of genus Ocimum L. Ecology and Evolution, 6: 7671-7682.
Lawrence, B.M. 1992. Chemical components of Labiatae oils and their exploitation. In: Harleyand, R.M. and Reynolds, T. (Eds). Advances in Labiate Science (pp. 399 - 436), Royal Botanic Gardens, Kew, UK, 399 - 436.
Leitch, I.J. and Bennett, M.D. 2004. Genome downsizing in polyploid plants. Biological Journal of the Linnean Society, 82: 651-663.
Lekhapan, P., Anamthawat-Jónsson, K. and Chokchaichamnankit, P. 2019. Chromosome evolution based on variation in chromosome number and chiasma frequency in the genus Ocimum L. from Thailand. Cytologia, 84: 199-206.
Levan, A., Fredga, K. and Sandberg, P. 1964. Nomenclature for centromeric position on chromosomes. Hereditas, 52: 201-220.
Lowry, D.B. and Willis, J.H. 2010. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLOS Biology, 8: e1000500. doi:1000510.1001371/journal.pbio.1000500.
Lysák, M.A. and Schubert, I. 2013. Mechanisms of chromosome rearrangements. In: Greilhuber, J., Dolezel, J. and Wendel, J.F. (Eds). Plant genome diversity: Physical structure, behaviour and evolution of plant genomes (pp. 137-147), Springer Vienna, Vienna, 137-147.
Mandáková, T., Hloušková, P., Koch, M.A. and Lysak, M.A. 2020. Genome evolution in Arabideae was marked by frequent centromere repositioning. The Plant Cell, 32: 650.
Mandáková, T. and Lysak, M.A. 2018. Post-polyploid diploidization and diversification through dysploid changes. Current Opinion in Plant Biology, 42: 55-65.
Mehra, P.N. and Gill, L.S. 1972. Cytology of West Himalayan Labiate: Tribe Ocimoideae. Cytologia, 37: 53-57.
Morton, J.K. 1962. Cytotaxonomic studies on the West African Labiatae. Journal of the Linnean Society of London, Botany, 58: 231-283.
Mukherjee, M. and Datta, A.K. 2006. Secondary chromosome associations in Ocimum spp. Cytologia, 71: 149-152.
Mukherjee, M., Datta, A.K. and Maiti, G.G. 2005. Chromosome number variation in Ocimum basilicum L. Cytologia, 70: 455-458.
Nathewet, P., Hummer, K.E., Yanagi, T., Iwatsubo, Y. and Sone, K. 2010. Karyotype analysis in octoploid and decaploid wild strawberries in Fragaria (Rosaceae). Cytologia, 75: 277-288.
Özer, I., Tamkoç, A. and Uysal, T. 2018. Determination of chromosome structures of genotypes of perennial ryegrass (Lolium perenne L.) selected from natural vegetation of Turkey. Cytologia, 83: 415-419.
Paton, A., Harley, R.M. and Harley, M.M. 1999. Ocimum-an overview of relationships and classification. In: Hiltunen, R. and Holm, Y. (Eds). Basil: The genus Ocimum (pp. 1-38), Harwood Academic, Amsterdam, 1-38.
Paton, A. and Putievsky, E. 1996. Taxonomic problems and cytotaxonomic relationships between and within varieties of Ocimum basilicum and related species (Labiatae). Kew Bulletin, 51: 509-524.
Paton, A.J., Springate, D., Suddee, S., Otieno, D., Grayer, R.J., Harley, M.M., Willis, F., Simmonds, M.S.J., Powell, M.P. and Savolainen, V. 2004. Phylogeny and evolution of basils and allies (Ocimeae, Labiatae) based on three plastid DNA regions. Molecular Phylogenetics and Evolution, 31: 277-299.
Pederson, T. 2011. The nucleolus. Cold Spring Harbor Perspectives in Biology, 3: doi:10.1101/cshperspect.a000638.
Pushpangadan, P. and Sobti, S.N. 1982. Cytogenetical studies in the genus Ocimum I. Origin of O. americanum, cytotaxonomical and experimental proof. Cytologia, 47: 575-583.
Pyne, R.M., Honig, J.A., Vaiciunas, J., Wyenandt, C.A. and Simon, J.E. 2018. Population structure, genetic diversity and downy mildew resistance among Ocimum species germplasm. BMC Plant Biology, 18: 69. doi:10.1186/s12870-12018-11284-12877
Rewers, M. and Jedrzejczyk, I. 2016. Genetic characterization of Ocimum genus using flow cytometry and inter-simple sequence repeat markers. Industrial Crops and Products, 91: 142-151.
Ryding, O. 1994. Notes on the sweet basil and its wild relatives (lamiaceae). Economic Botany, 48: 65-67.
Schranz, M.E., Lysak, M.A. and Mitchell-Olds, T. 2006. The ABC's of comparative genomics in the Brassicaceae: Building blocks of crucifer genomes. Trends in Plant Science, 11: 535-542.
Schubert, I. and Vu, G.T.H. 2016. Genome stability and evolution: Attempting a holistic view. Trends in Plant Science, 21: 749-757.
Smitinand, T. 2014. Thai plant names "Tem Smitinand" revised edition 2014. In: Bangkok Forest Herbarium, Bangkok, 826 pp.
Soltis, D.E., Visger, C.J., Marchant, D.B. and Soltis, P.S. 2016. Polyploidy: Pitfalls and paths to a paradigm. American Journal of Botany, 103: 1146-1166.
Stebbins, G.L. 1971. Chromosomal evolution in higher plants. In: Edward Arnold, London, 216 pp.
Suddee, S., Paton, A.J. and Parnell, J.A.N. 2005. Taxonomic revision of tribe Ocimeae Dumort. (Lamiaceae) in continental South East Asia III. Ociminae. Kew Bulletin, 60: 3-75.
Tapia, F., Gomez, S. and Mercado-Ruaro, P. 2018. Differences in karyotypes between two populations of Crotalaria incana from Mexico. Cytologia, 83: 431-435.
Thoppil, J.E. and Jose, J. 1994. Infraspecific genetic control of major essential oil constituents in Ocimum basilicum Linn. The Nucleus, 37: 30-33.
Vieira, R.F. and Simon, J.E. 2000. Chemical characterization of basil (Ocimum spp.) found in the markets and used in traditional medicine in Brazil. Economic Botany, 54: 207-216.
Vij, S.P. and Kashyap, S.K. 1976. Cytological studies in some North Indian Labiatae. Cytologia, 41: 713-719.
Zarco, C.R. 1986. A new method for estimating karyotype asymmetry. Taxon, 35: 526-530.
Downloads
Published
How to Cite
Issue
Section
License
Chulalongkorn University. All rights reserved. No part of this publication may be reproduced, translated, stored in a retrieval system, or transmitted in any form or by any means, electronic, mechanical, photocopying, recording or otherwise, without prior written permission of the publisher











