Taxonomic Status of Fowlea tytleri (Blyth, 1863) from the Andaman Islands
DOI:
https://doi.org/10.58837/tnh.21.3.253426Keywords:
Biodiversity studies, mtDNA, reptiles, Southeast Asia, XenochrophisAbstract
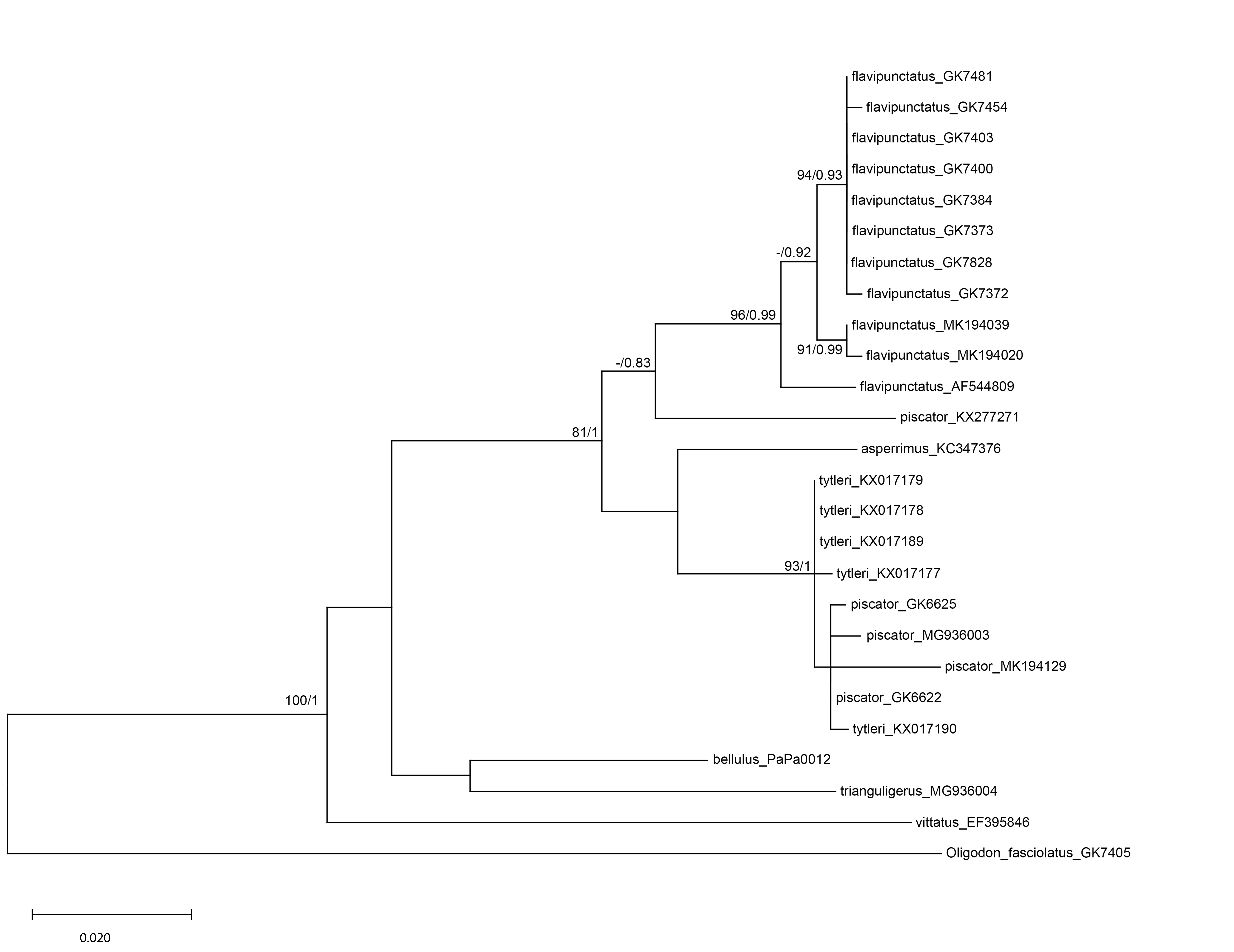
The analysis of mtDNA data (16S ribosomal RNA) shows that the Fowlea (formerly Xenochrophis) of the Andaman islands, formerly referred to as Fowlea (or Xenochrophis) tytleri are conspecific with F. piscator (Schneider, 1799). Specimens from the Andaman islands form a mixed clade together with individuals from the mainland and differ from the latter by genetic distances of 0.2–0.5% (except for 1.7% from a single mainland specimen that also differs from other mainland samples of F. piscator by 1.4–1.5% genetic distance). The genetic distances between individuals of F. piscator and its supposed sister taxon F. flavipunctatus vary from 4.8 to 7.3%. In pholidosis, F. tytleri and F. piscator are essentially identical. Based on the evidence from mtDNA and morphological data (the latter taken from the literature), we formally place Fowlea tytleri (Blyth, 1863) in the synonymy of F. piscator (Schneider, 1799).
References
Bandopadhyay, P.C. and Carter, A. 2017. Chapter 6. Geological framework of the Andaman-Nicobar islands. Geological Society, London, Memoirs, 47: 75–93.
Biswas, S. 1984. Some notes on the reptiles of the Andaman and Nicobar Islands. Journal of the Bombay Natural History, 81: 476–481.
Blyth, E. 1863. Report of the Curator, Zoological Department. Journal of the Asiatic Society of Bengal, 32: 79–90.
Burnham, K.P. and Anderson, D.R. 2002. Model Selection and Multimodel Inference. A Practical Information-Theoretic Approach. Springer-Verlag New York Inc, New York, NY.
Edgar, R.C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC bioinformatics, 5: 1–19.
Gelman, A. and Rubin, D.B. 1992. Inference from iterative simulation using multiple sequences. Statistical Science, 7: 457–472.
Glez-Peña, D., Gómez-Blanco, D., Reboiro-Jato, M., Fdez-Riverola, F. and Posada, D. 2010. ALTER: program-oriented conversion of DNA and protein alignments. Nucleic Acids Research, 38: 14-18.
Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W. and Gascuel, O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic biology, 59: 307–321.
Ivanova, N.V., Waard, J. de and Hebert, P.D. 2006. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Molecular Ecology Notes, 6: 998–1002.
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., Buxton, S., Cooper, A., Markowitz, S., Duran, C., Thierer, T., Ashton, B., Meintjes, P. and Drummond, A. 2012. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28: 1647–1649.
Khan, M.S. 2002. Die Schlangen Pakistans. Edition Chimaira, Frankfurt, 265 pp.
Kumar, S., Stecher, G. and Tamura, K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Molecular Biology and Evolution, 33: 1870–1874.
Lanfear, R., Frandsen, P.B., Wright, A.M., Senfeld, T. and Calcott, B. 2017. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution, 34: 772–773.
Minh, B.Q., Nguyen, M.A.T. and Haeseler, A. von 2013. Ultrafast approximation for phylogenetic bootstrap. Molecular Biology and Evolution, 30: 1188–1195.
Mohan, A.V., Swamy, P. and Shanker, K. 2018. Population structure in the Andaman keelback, Xenochrophis tytleri: geographical distance and oceanic barriers to dispersal influence genetic divergence on the Andaman archipelago. PeerJ: 6:e5752.
Purkayastha, J., Kalita, J., Brahma, R.K., Doley, R. and Das, M. 2018. A review of the relationships of Xenochrophis cerasogaster Cantor, 1839 (Serpentes: Colubridae) to its congeners. Zootaxa, 4514: 126–136.
Pyron, R.A., Burbrink, F.T. and Wiens, J.J. 2013. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evolutionary Biology, 13: 93.
Ronquist, F., Teslenko, M., Mark, P., Ayres, D., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M. and Huelsenbeck, J. 2012. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Systematic biology, 61: 539–542.
Schleich, H.H. and Kästle, W. 2002. Amphibians and Reptiles of Nepal. A.R.G. Gantner Verlag, Ruggell, 1201 pp.
Simpson, G.G. 1951. The species concept. Evolution: 285–298.
Smith, M.A. 1943. The Fauna of British India, Ceylon and Burma, including the whole of the Indo-Chinese Subregion. Reptilia and Amphibia. Vol. III. - Serpentes. Taylor & Francis, London, 583 pp.
Trifinopoulos, J., Nguyen, L.-T., Haeseler, A. von and Minh, B.Q. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research, 44: W232-W235.
Vogel, G. and David, P. 2006. On the taxonomy of the Xenochrophis piscator complex (Serpentes, Natricidae). In: Vences, M., Köhler, J., Ziegler, T. and Böhme, W. (Eds.). Herpetologia Bonnensis II. Proceedings of the 13th Congress of the Societas Europaea Herpetologica, Bonn, pp. 241–246.
Vogel, G. and David, P. 2012. A revision of the species group of Xenochrophis piscator (Schneider, 1799) (Squamata: Natricidae). Zootaxa, 3473: 1–60.
Whitaker, R. and Captain, A. 2004. Snakes of India. The field guide. Draco Books, Chennai, 479 pp.
Wiley, E.O. 1978. The Evolutionary Species Concept Reconsidered. Systematic biology, 27: 17–26.
Downloads
Published
How to Cite
Issue
Section
License
Chulalongkorn University. All rights reserved. No part of this publication may be reproduced, translated, stored in a retrieval system, or transmitted in any form or by any means, electronic, mechanical, photocopying, recording or otherwise, without prior written permission of the publisher











