Diversity and Distribution of Jellyfish Polyps Along Coastal Areas of Chonburi and Rayong Provinces, Thailand
DOI:
https://doi.org/10.58837/tnh.23.1.258166Keywords:
COI, DNA, morphology, polyp, jellyfishAbstract
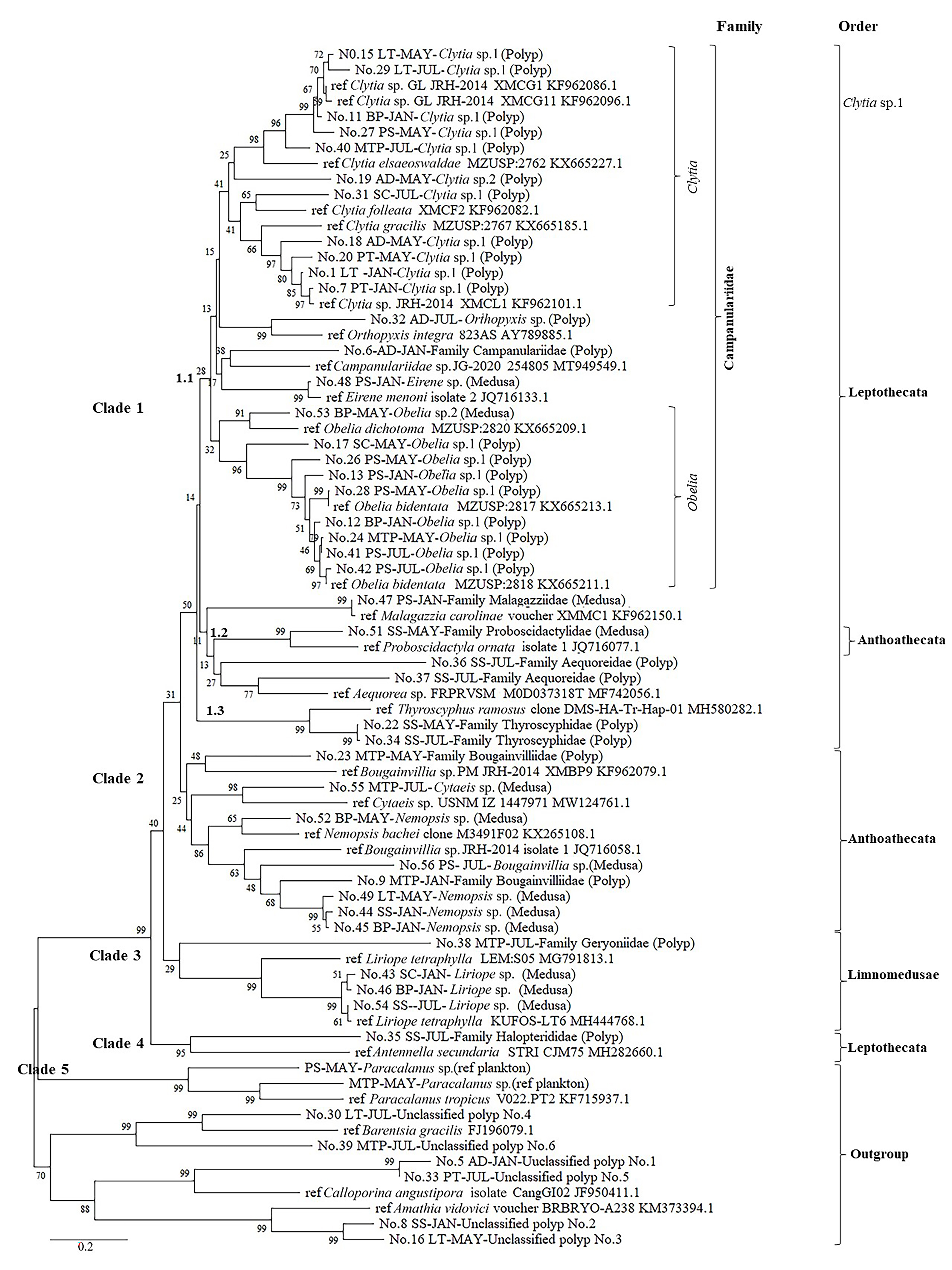
Jellyfish polyps can be difficult to identify based on their morphology due to a lack of precise references in Thailand, yet species identification is an important step for management of this marine resource. Here, we pursued a dual approach, morphology and DNA barcoding, to describe the diversity and distribution of jellyfish polyps in two coastal provinces that have various anthropogenic activities e.g., tourism, marine transportations, industrial estate, local fisheries, and aquacultures, that effect on water qualities or provide substrates for polyp settlement. Jellyfish polyps were collected in January, May, and July 2019 to represent the Northeast Monsoon, pre-Southwest Monsoon, and Southwest Monsoon, respectively, from eight stations along coastal areas from Chonburi and Rayong Provinces, eastern Thailand. The jellyfish polyps were sampled from substrates e.g., rocks, ropes, and shells, at sampling sites by scuba diving. Three genera of polyps were identified according to their morphology, while nine genera of jellyfish polyps were identified by their COI gene sequences from 29 individuals. Polyps of the genus Clytia were recorded during the sampling periods at most sampling sites, while the genus Obelia was found mainly at Rayong Province, when identified by both morphological and molecular approaches. These results can be used as part of a suitable management plan about jellyfish issues in Thailand.
References
Adamík P., Gallager S.M., Horgan E., Madin L.P., McGillis W.R., Govindarajan A. and Alatalo P. 2006. Effects of turbulence on the feeding rate of a pelagic predator: the planktonic hydroid Clytia gracilis. Journal of Experimental Marine Biology Ecology, 333: 159–165.
Andújar, C., Arribas, P., Yu, D.W., Vogler, A.P., and Emerson, B.C. 2018. Why the COI barcode should be the community DNA metabarcode for the Metazoa. Molecular Ecology, 27(20): 3968-3975.
Baek, S.Y., Jang, K.H., Choi, E.H., Ryu, S.H., Kim, S.K., Lee, J.H., Lim, Y.J., Lee, J., Jun, J., Kwak, M., Lee, Y.S., Hwang, J.S., Venmathi Maran, B.A., Chang, C.Y., Kim, I.H., and Hwang, U.W. 2016. DNA Barcoding of Metazoan Zooplankton Copepods from South Korea. PloS one, 11(7): e0157307.Bendezu, I.F., Slater, J.W. and Carney, B.F. 2005. Identification of Mytilus spp. and Pecten maximus in Irish Waters by Standard PCR of the 18S rDNA Gene and Multiplex PCR of the 16S rDNA Gene. Marine Biotechnology, 7: 687–696.
Bingpeng, X., Heshan, L., Zhilan, Z., Chunguang, W., Yanguo, W., Jianjun, W. 2018) DNA barcoding for identification of fish species in the Taiwan Strait. PLoS ONE 13(6): e0198109.
Boero, F. 2013. Review of jellyfish blooms in the Mediterranean and Black Sea. Studies and Reviews. General Fisheries Commission for the Mediterranean. No. 92. FAO, Rome, 53 pp.
Boero F., Di Camillo C. and Gravili C. 2005. Aquatic invasions: phantom aliens in Mediterranean waters. MarBEF Newsletter, 3: 21–22.
Brotz, L., Cheung, W.W.L., Kleisner, K., Pakhomov, E. and Pauly, D. 2012. Increasing jellyfish populations: trends in Large Marine Ecosystems. Hydrobiologia, 690: 3–20.
Brotz, L. and Pauly, D. 2012. Jellyfish populations in the Mediterranean Sea. Acta Adriatica, 53(2): 211–230.
Bucklin, A., Batta-Lona, P.G., Questel, J.M., Wiebe, P.H., Richardson, D.E., Copley, N.J. and O’Brien, T.D. 2022. COI Metabarcoding of Zooplankton Species Diversity for Time-Series Monitoring of the NW Atlantic Continental Shelf. Frontiers in Marine Science, 9: 867893.
Bucklin, A., Ortman, B.D., Jennings, R.M., Nigro, L.M., Sweetman, C.J., Copley, N.J., Wiebe, P.H. 2010. A “Rosetta Stone” for zooplankton: DNA barcode analysis of holozooplankton diversity of the Sargasso Sea (NW Atlantic Ocean). Deep-Sea Res II, 57: 2234–2247.
Bucklin, A., Peijnenburg, K.T.C.A., Kosobokova, K.N., O’Brien, T.D., Blanco-Bercial, L., Cornils, A., Falkenhaug, T., Hopcroft, R.R., Hosia, A., Laakmann, S., Li, C., Martell, L., Questel, J.M., Wall-Palmer, D., Wang, M., Wiebe, P.H. and Weydmann-Zwolicka, A. 2021. Toward a global reference database of COI barcodes for marine zooplankton. Marine Biology, 168: 78.
Bucklin, A., Steinke, D. and Blanco-Bercial, L. 2011. DNA Barcoding of Marine Metazoa. Annual Review of Marine Science, 3: 471–508.
Buranapratheprat, A. 2008. Circulation in the Upper Gulf of Thailand: A review. Burapha Science Journal. 13(1), 75–83.
Calder, D.R. 1991. Shallow-water hydroids of Bermuda: the Thecatae, exclusive of Plumularioidea. Royal Ontario Museum, Toronto, Life Science Contributions, 154: 1–140.
Calder, D.R. 2012. On a collection of hydroids (Cnidaria, Hydrozoa, Hydroidolina) from the west coast of Sweden, with a checklist of species from the region. Zootaxa, 3171: 1–77.
Chae, J., Seo, Y., Yu, W.B., Yoon, W.D., Lee, H.E., Chang, S.J. and Ki, J.S. 2018. Comprehensive analysis of the jellyfish Chrysaora pacifica (Goette, 1886) (Semaeostomeae: Pelagiidae) with description of the complete rDNA sequence. Zoological Studies, 57: 51.
Choosri, S., Luangoon, N., Charoendee, W., Taweedetch, S. and Phuangsanthia, W. 2016. Life cycle of the jellyfish Catostylus sp. Final report, The Institute of Marine Science, Burapha University, Chonburi, 42 pp.
Condon R.H., Duarte, C.M., Pitt, K.A., Robinson, K.L., Lucas, C.H., Sutherland, K.R., Mianzan, H.W., Bogeberg, M., Purcell, J.E., Decker, M.B., Uye, S., Madin, L.P., Brodeur, R.D., Haddock, S.H.D., Malej, A., Parry, G.D., Eriksen, E., Quiñones, J., Acha, M., Harvey, M., Arthur, J.M. and Graham, W.M. 2013. Recurrent jellyfish blooms are a consequence of global oscillations. Proceedings of the National Academy of Sciences of the United States of America, 110(3): 1000–1005.
Cornelius, P.F.S. 1975. The hydroid species of Obelia (Coelenterata, Hydrozoa: Campanulariidae), with notes on the medusa stage. Bulletin of the British Museum, Zoology 28: 251–293.
Cornelius, P.F.S 1982. Hydroids and medusae of the family Campanulariidae recorded from the eastern North Atlantic, with a world synopsis of genera. Bulletin of the British Museum, Zoology 42: 37–148.
Costello, J.H. and Colin, S.P. 2002. Prey resource use by coexistent hydromedusae from Friday Harbor, Washington. Limnology and Oceanography, 47(4): 934–942.
Di Camillo, C.G., Bavestrello, G., Valisano, L. and Puce, S. 2008. Spatial and temporal distribution in a tropical hydroid assemblage. Journal of the Marine Biological Association of the United Kingdom, 88(8): 1589–1599.
Dong, Z., Liu, D. and Keesing, J.K. 2010. Jellyfish blooms in China: Dominant species, causes and consequences. Marine Pollution Bulletin, 60: 954–963.
Ershova, E.A., Wangensteen, O.S., Descoteaux, R., Barth-Jensen, C. and Prabel, K. 2021. Metabarcoding as a quantitative tool for estimating biodiversity and relative biomass of marine zooplankton. ICES Journal of Marine Science, 78(9): 3342–3355.
Folmer, O., Black, M., Hoeh, W., Lutz, R. and Vrijenhoek, R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular marine biology and biotechnology, 3(5): 294–299.
Fulton, R.S. and Wear, R.G. 1985. Predatory feeding of the hydromedusae Obelia geniculata and Phialella quadrata. Marine Biology, 87: 47–54.
Galan, G.L., Mendez, N.P. and dela Cruz, R.Y. 2018. DNA barcoding of three selected gastropod species using cytochrome oxidase (COI) gene. Annales of West University of Timisoara, Series of Biology, 21(1): 93–102.
Ge, Y., Xia, C., Wang, J., Zhang, X., Ma, X., and Zhou, Q. 2021. The efficacy of DNA barcoding in the classification, genetic differentiation, and biodiversity assessment of benthic macroinvertebrates. Ecology and evolution, 11(10): 5669–5681.
Gershwin, L., Condie, S.A., Mansbridge, J.V. and Richardson, A.J. 2014 Dangerous jellyfish blooms are predictable. Journal of The Royal Society Interface, 11: 20131168.
Gravili, C, Vito, D.D, Camillo, C.G.D., Martell, L., Piraino, S., Boero, F., 2015. The non-Siphonophoran Hydrozoa (Cnidaria) of Salento, Italy with notes on their life-cycles: an illustrated guide. Zootaxa, 3908 (1): 001–187.
Green, T.J., Wolfenden, D.C.C. and Sneddon, L.U. 2018. An investigation on the impact of substrate type, temperature, and iodine on moon jellyfish production. Zoo Biology, 37: 434–439.
Hall, T.A. 1999. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95-98.
Hansson, L.J., Moeslund, O., Kiørboe, T. and Riisgård, H.U. 2005. Clearance rates of jellyfish and their potential predation impact on zooplankton and fish larvae in a neritic ecosystem (Limfjorden, Denmark). Marine Ecology Progress Series, 304: 117–131.
He, J., Zheng, L., Zhang, W., Lin, Y. and Cao, W. 2015. Morphology and molecular analyses of a new Clytia species (Cnidaria: Hydrozoa: Campanulariidae) from the East China Sea. Journal of the Marine Biological Association of the United Kingdom, 95(2): 289–300.
Hebert, P.D., Cywinska, A., Ball, S.L. and deWaard, J.R. 2003. Biological identifications through DNA barcodes. Proceedings. Biological sciences, 270(1512): 313–321.
Holst, S. and Jarms, G. 2007. Substrate choice and settlement preferences of planula larvae of five Scyphozoa (Cnidaria) from German Bight, North Sea. Marine Biology, 151: 863–871.
Hoover, R.A., and Purcell, J.E. 2009. Substrate preferences of scyphozoan Aurelia labiata polyps among common dock-building materials. Hydrobiologia, 616(1): 259–267.
Huang, D., Meier, R., Todd, P.A. and Chou, L.M. 2008. Slow Mitochondrial COI Sequence Evolution at the Base of the Metazoan Tree and Its Implications for DNA Barcoding. Journal of Molecular Evolution, 66: 167–174.
Hwai, A.T.S., Peng, C.T.C., Nilamani, N., and Yasin, Z. 2019. Harmful Jellyfish Country Report in Western Pacific. Centre for Marine and Coastal Studies, Universiti Sains Malaysia, Penang, Malaysia. 61 pp.
Jitrapat, H. 2018. Abundance and diversity of gelatinous zooplankton in the Inner Gulf of Thailand. Master thesis, Department of Marine Science, Faculty of Science, Chulalongkorn University, 145 pp.
Kitajima, S., Iguchi, N., Honda, N., Watanabe, T. and Katoh, O. 2015. Distribution of Nemopilema nomurai in the southwestern Sea of Japan related to meandering of the Tsushima Warm Current. Journal of Oceanography, 71: 287–296.
Klomtong, P., Phasuk, Y., and Duangjinda, M. 2016. Animal Species Identification through High Resolution Melting Real Time PCR (HRM) of the Mitochondrial 16S rRNA Gene. Annals of Animal Science, 16(2): 415–424.
Kubota, S. 1978. The Life-History of Clytia edwardsi (Hydrozoa; Campanulariidae) in Hokkaido, Japan. Journal of the Faculty of Science, Hokkaido University. Series 6, Zoology, 21(3): 317–354.
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35(6): 1547–1549.
Laakmann, S. and Holst, S. 2014. Emphasizing the diversity of North Sea hydromedusae by combined morphological and molecular methods. Journal of Plankton Research, 36(1): 64–76.
Lianming, Z., Jinru, H., Yuanshao, L., Wenqing, C. and Wenjing, Z. 2014. 16S rRNA is a better choice than COI for DNA barcoding hydrozoans in the coastal waters of China. Acta Oceanologica Sinica, 33(4): 55–76.
Lindsay, D.J., Grossmann, M.M., Nishikawa, J., Bentlage, B., and Collins, A.G. 2015. DNA barcoding of pelagic cnidarians: current status and future prospects. Bulletin of the Plankton Society of Japan, 62(1): 39–43.
Lindner, A. and Migotto, A.E. 2002. The life cycle of Clytia linearis and Clytia noliformis: metagenic campanulariids (Cnidaria: Hydrozoa) with contrasting polyp and medusa stages. Journal of the Marine Biological Association of the United Kingdom, 82: 541–553.
Lo, W.T., Purcell, J.E., Hung, J.J., Su, H.M. and Hsu, P.K. 2008. Enhancement of jellyfish (Aurelia aurita) populations by extensive aquaculture rafts in a coastal lagoon in Taiwan. ICES Journal of Marine Science, 65: 453–461.
Lucas, C.H., Graham, W. and Widmer, C.L. 2012. Jellyfish life histories: role of polyps in forming and maintaining scyphomedusa populations. In: Lesser, M. (Ed.) Advances in Marine Biology, Vol.63, Academic Press, 133–196.
Lucas, C.H., Williams, D.W., Wiliams, J.A. and Sheader, M. 1995. Seasonal dynamics and production of the hydromedusan Clytia hemisphaerica (Hydromedusa: Leptomedusa) in Southampton water. Estuaries, 18: 362–372.
Macher, J.N., Wideman, J.G., Girard, E.B., Langerak, A., Duijm, E., Jompa, J., Sadekov, A., Vos, R., Wissels, R., and Renema, W. 2021. First report of mitochondrial COI in foraminifera and implications for DNA barcoding. Scientific reports, 11(1): 22165.
Mariac, C., Vigouroux, Y., Duponchelle, F., García-Dávila, C., Nunez, J., Desmarais, E., and Renno, J.F. 2018. Metabarcoding by capture using a single COI probe (MCSP) to identify and quantify fish species in ichthyoplankton swarms. PLOS ONE, 13(9): e0202976.
Marine and Coastal Resources Research and Development Institute. 2015a. Thai Marine and Coastal Resources Scriptures 2015. Department of Marine and Coastal Resources, Ministry of Natural Resources and Environment, Bangkok, 246 pp.
Marine and Coastal Resources Research and Development Institute. 2015b. Guide to the diversity of jellyfish in the waters Thailand. Department of Marine and Coastal Resources, Ministry of Natural Resources and Environment, Bangkok, 127 pp.
Mendoza-Becerril, M.A., Estrada-GonzÁlez, M.C., Mazariegos-Villarreal, A., Restrepo-AvendaÑo, L., Villar-BeltrÁn, R.D., AgÜero, J., and Cunha, A.F. 2020. Taxonomy and diversity of Hydrozoa (Cnidaria, Medusozoa) of La Paz Bay, Gulf of California. Zootaxa, 4808(1): 1–37.
Miglietta, M.P., Rossi, M. and Collin, R. 2008. Hydromedusa blooms and upwelling events in the Bay of Panama, Tropical East Pacific. Journal of Plankton Research, 30(7): 783–793.
Moura, C.J., Harris, D.J., Cunha, M.R. and Rogers, A.D. 2008. DNA barcoding reveals cryptic diversity in marine hydroids (Cnidaria, Hydrozoa) from coastal and deep-sea environments. Zoologica Scripta, 37: 93–108.
Moura, C.J., Lessios, H., Cortés, J., Nizinski, M.S., Reed, J., Santos, R.S., and Collins, A.G. 2018. Hundreds of genetic barcodes of the species-rich hydroid superfamily Plumularioidea (Cnidaria, Medusozoa) provide a guide toward more reliable taxonomy. Scientific reports, 8(1): 17986.
Muhammad, B.L., Seo, Y. and Ki, J.S. 2021 Evaluation ofthe complete nuclear rDNA unit sequence of the jellyfish Cyanea nozakii Kishinouye (Scyphozoa: Semaeostomeae) for molecular discrimination, Animal Cells and Systems, 25:6, 424–433.
National Center for Biotechnology Information (NCBI)[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988] – [cited 2020 Sep 13]. Available from: https://www.ncbi.nlm.nih.gov/
Ortman, B.D., Bucklin, A., Pages, F. and Youngbluth, M. 2010. DNA Barcoding the Medusozoa using mtCOI. Deep-Sea Research Part II, 57: 2148–2156.
Pappalardo, P., Collins, A.G., Pagenkopp Lohan, K.M., Hanson, K.M., Truskey, S.B., Jaeckle, W., Ames, C.L., Goodheart, J.A., Bush, S.L., Biancani, L.M., Strong, E.E., Vecchione, M., Harasewych, M.G., Reed, K., Lin, C., Hartil, E.C., Whelpley, J., Blumberg, J., Matterson, K., Redmond, N.E., Becker, A., Boyle, M.J., and Osborn, K.J. 2021. The role of taxonomic expertise in interpretation of metabarcoding studies. ICES Journal of Marine Science, doi:10.1093/icesjms/fsab082.
Parracho, T., and Morais, Z. 2015. Catostylus tagi: partial rDNA sequencing and characterisation of nematocyte structures using two improvements in jellyfish sample preparation. Journal of Venomous Animals and Toxins Including Tropical Diseases, 21: 40.
Patithanarak, T., Luang-on, N., Manhaphol, A., Taleb, S. and W. 2014. Molecular Identification of Colorful Jellyfish along the Coast of Trat Province. The 4th Marine Science Conference, Prince of Songkhla University, Songkhla, 287–296.
Phongphattarawat, S. 2013. Species diversity and abundance of zooplankton Class Hydrozoa (Cnidaria: Hydrozoa) in the Inner Gulf of Thailand. Senior project, Department of Biology, Faculty of Science, Chulalongkorn University, 79 pp.
Phuangsanthia, W., Choosri, S., Chunam, W. and Sripanomyom, J. 2018. Life Cycle and Development of Upside-Down Jellyfish, Cassiopea andromeda (Forsskal, 1775) in Laboratory Conditions. Burapha Science Journal, 23(2): 1178–1187.
Phuangsanthia, W., Choosri, S., Fakrajang, S. and Muthuwan, V. 2020. Life cycle of the edible jellyfish Rhopilema hispidum (Vanhöffen, 1888). Khon Kaen Agriculture Journal, 48(Suppl.1): 907–916.
Pinto, C.D.C., 2021. The effect of temperature and substrates on polyp settlement of the spotted jellyfish, Phyllorhiza punctata (Von Lendenfeld, 1884). Master thesis in Aquaculture, Politécnico de Leiria, Portugal. 30 pp.
Pitt, K.A., Lucas, C.H., Condon, R.H., Duarte, C.M., and Stewart-Koster, B. 2018. Claims That Anthropogenic Stressors Facilitate Jellyfish Blooms Have Been Amplified Beyond the Available Evidence: A Systematic Review. Frontiers in Marine Science, 5: 451.
Purcell, J.E., Uye, S. and Lo, W.T. 2007. Anthropogenic causes of jellyfish blooms and their direct consequences for humans: a review. Marine Ecology Progress Series, 350: 153–174.
Radulovici, A.E., Archambault, P. and Dufresne, F. 2010. DNA Barcodes for Marine Biodiversity: Moving Fast Forward? Diversity, 2: 450–472.
Ran, K., Li, Q., Qi, L., Li, W., and Kong, L. 2020. DNA barcoding for identification of marine gastropod species from Hainan Island, China. Fisheries Research, 225: 105504.
Richardson, A.J., Bakun, A., Hays, G.C. and Gibbons, M.J. 2009. The jellyfish joyride: causes, consequences and management responses to a more gelatinous future. Trends in Ecology and Evolution, 24(6): 312–322.
Ruijuan, L., Jie, X., Xuelei, Z. and Aungtonya, C. 2016. Genetic analysis of common venomous Cubozoa and Scyphozoa in Thailand waters. Haiyang Xuebao. 38(6): 51-61. (in Chinese with English abstract).
Sathirapongsasuti, N., Khonchom, K., Poonsawat, T., Pransilpa,M., Ongsara, S., Detsri, U., Bungbai, S., Lawanangkoon, S.-a., Pattanaporkrattana,W. and Trakulsrichai, S. 2021. Rapid and Accurate Species-Specific PCR for the Identification of Lethal Chironex Box Jellyfish in Thailand. International Journal of Environmental Research and Public Health, 18, 219.
Schuchert, P. 2003. Hydroids (Cnidaria, Hydrozoa) of the Danish expedition to the Kei Islands. Steenstrupia, 27(2): 137–256.
Schuchert, P. 2018. DNA barcoding of some Pandeidae species (Cnidaria, Hydrozoa, Anthoathecata). Revue Suisse De Zoologie, 125(1): 101–127.
Schuchert, P. 2020. World Hydrozoa Database, Flanders Marine Institute. Available from: http://www.Marinespecies. org/hydrozoa/, (13 August 2020).
Sojisuporn, P., Morimoto, A. and Yanagi, T. 2010. Seasonal variation of sea surface current in the Gulf of Thailand. Coastal Marine Science, 34(1): 91–102.
Song, F., Jianing, L., Song, S. and Fang, Z. 2017. Artificial substrates preference for proliferation and immigration in Aurelia aurita (s. 1.) polyps. Journal of Oceanology and Limnology, 35(1): 153–162.
Sun, S., Zhang, F., Li, C.L., Wang, S.W., Wang, M.X., Tao, Z.C., Wang, Y.T., Zhang, G.T. and Sun, X.X. 2015. Breeding place, population development and distribution pattern of the giant jellyfish Nemopilema nomurai (Scyphozoa: Rhizostomeae) in the Yellow Sea and East China Sea. Hydrobiologia, 754: 59–74.
Sutherland, K.R., Gemmell, B.J., Colin, S.P. and Costello, J.H. 2016. Prey capture by the cosmopolitan hydromedusae, Obelia spp., in the viscous regime. Limnology and Oceanography, 61: 2309–2317.
Toshino, S., Nishikawa, J., Srinui, K., Taleb, S. and Miyake, H. 2019. New records of two species of Cubozoa from Thailand. Plankton and Benthos Research, 14(3): 143–149.
Uye, S. 2008. Blooms of the giant jellyfish Nemopilema nomurai: a threat to the fisheries sustainability of the East Asian Marginal Seas. Plankton Benthos Research, 3 (Suppl.): 125–131.
Uye, S. 2010. Jellyfish Blooms in East Asian Coastal Seas: Loss of Productive Ecosystem. International Symposium on Integrated Coastal Management for Marine Biodiversity in Asia, Kyoto, Japan, 16–19.
van Walraven, L., Driessen, F., van Bleijswijk, J., Bol, A., Luttikhuizen, P.C., Coolen, J.W., Bos, O.G., Gittenberger, A., Schrieken, N., Langenberg, V.T., and van der Veer, H.W. 2016. Where are the polyps? Molecular identification, distribution and population differentiation of Aurelia aurita jellyfish polyps in the southern North Sea area. Marine biology, 163: 172.
van Walraven, L., van Bleijswijk, J., van der Veer, H.W. 2020. Here are the polyps: in situ observations of jellyfish polyps and podocysts on bivalve shells. PeerJ 8: e9260.
Wilson, E. 2002. Obelia bidentata A hydroid. In Tyler-Walters, H. and Hiscock, K. (eds.) Marine Life Information Network: Biology and Sensitivity Key Information Reviews, Plymouth: Marine Biological Association of the United Kingdom. Available from: http://www.marlin.ac.uk/species/detail/1270/, (10 August 2020)
Wuttichareonmongkol, N. 2004. Diversity and Abundance of Planktonic Hydrozoan, Class Hydrozoa, in the Upper Gulf of Thailand. Master Thesis, Graduate School, Kasetsart University, Bangkok, 265 pp.
Zelickman, E.A. 1972. Distribution and Ecology of the Pelagic Hydromedusae, Siphonophores and Ctenophores of the Barents Sea, Based on Perennial Plankton Collections. Marine Biology, 17: 256–264.
Zhao, L., Zhang, X., Xu, M., Mao, Y. and Huang, Y. 2021. DNA metabarcoding of zooplankton communities: species diversity and seasonal variation revealed by 18S rRNA and COI. PeerJ, 9: e11057.
Zheng, L., He, J., Lin, Y., Cao, W., and Zhang, W. 2014. 16S rRNA is a better choice than COI for DNA barcoding hydrozoans in the coastal waters of China. Acta Oceanologica Sinica, 33(4): 55–76.
Zhen-zu, X., Jia-qi, H., Mao, L., Donghui, G. and Chunguang, W. 2014. The superclass hydrozoa of the phylum Cnidaria in China, Vol.2, China Ocean Press, Beijing, 945 pp.
Zhou, K., Zheng, L., He, J., Lin, Y., Cao, W. and Zhang, W. 2013. Detection of a new Clytia species (Cnidaria: Hydrozoa: Campanulariidae) with DNA barcoding and life cycle analyses. Journal of the Marine Biological Association of the United Kingdom, 93(8): 2075–2088.
Zou, R., Liang, C., Dai, M., Wang, X., Zhang, X. and Song, Z. 2020. DNA barcoding and phylogenetic analysis of bagrid catfish in China based on mitochondrial COI gene, Mitochondrial DNA Part A, 31(2): 73-80.
Downloads
Published
How to Cite
Issue
Section
Categories
License
Chulalongkorn University. All rights reserved. No part of this publication may be reproduced, translated, stored in a retrieval system, or transmitted in any form or by any means, electronic, mechanical, photocopying, recording or otherwise, without prior written permission of the publisher











