Proteomic analysis of cell-cell signaling alteration in rat frontal cortex following methamphetamine exposure
Keywords:
Methamphetamine, Frontal cortex, cell-cell signaling, ProteomicsAbstract
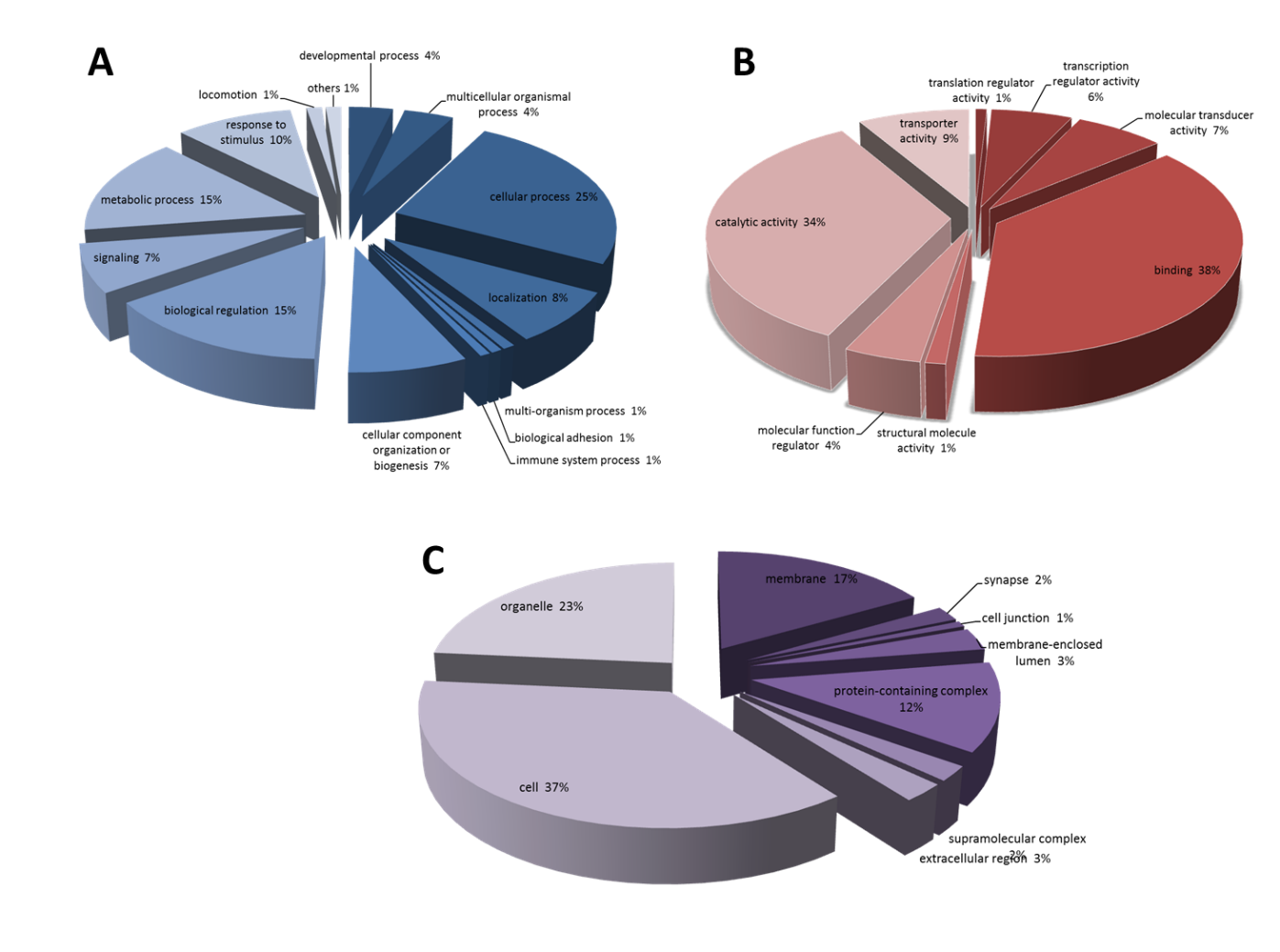
Methamphetamine (METH) is an addictive psychostimulant with potent effects on the central nervous system (CNS). Prolonged use of METH can impair brain structures and functions, especially the frontal cortex, a key brain involved in behavioral and cognitive functions. Moreover, METH has been reported change a number of proteins in neurotransmitter systems as well as proteins related to synaptic functions. Therefore, the objective of this study was to use the proteomic approach to investigate the differential expression of proteins related to synaptic function, including cell-cell signaling, in frontal cortex after METH administration. 20 male Sprague-Dawley rats were divided into 2 groups of control and METH; the rats were treated with saline and escalating binge dose of METH (0.1 to 4 mg/kg of METH (3 times /day), for 14 days and binge dose, 6 mg/kg (4 times /day) at day 15), respectively. The proteins in rat frontal cortex were investigated by proteomics technique. The results showed that there were 1,312 differentially expressed proteins in the frontal cortex of control and METH rats. Fifty-eight proteins were grouped in cell-cell signaling proteins. Thirty-six proteins were down-regulated and twenty-two proteins were up-regulated following METH administration. Furthermore, METH-interacted cell signaling proteins were mostly involved in neurotransmitter systems, 10 proteins in glutamatergic system, including 5 proteins in GABAergic system and 6 proteins in acetylcholine system. The results suggested that METH administration affects changes of proteins related in cell-cell signaling of the brain. These effects may implicate in METH-induced neurotoxicity. Studying in the differentially expressed protein by proteomic approach provides potential proteins related to METH-induced neurotoxicity
References
Vearrier D, Greenberg MI, Miller SN, Okaneku JT, Haggerty DA. Methamphetamine: history, pathophysiology, adverse health effects, current trends, and hazards associated with the clandestine manufacture of methamphetamine. Dis Mon, 2012;58(2):38-89.
Cruickshank CC, Dyer KR. A review of the clinical pharmacology of methamphetamine. Addiction, 2009;104(7): 1085-99.
Jan RK, Kydd RR, Russell BR. Functional and structural brain changes associated with methamphetamine abuse. Brain Sci, 2012;2(4):434-482.
Volkow ND, Wang GJ, Tomasi D, Baler RD. Unbalanced neuronal circuits in addiction. Curr Opin Neurobiol, 2013;23(4):639-48.
Kamei H, Nagai T, Nakano H, Togan Y, Takayanagi M, Takahashi K, et al. Repeated methamphetamine treatment impairs recognition memory through a failure of novelty-induced ERK1/2 activation in the prefrontal cortex of mice. Biolpsychiatry, 2006;59:75–84.
Ghasemzadeh MB, Vasudevan P, Giles C, Purgianto A, Seubert C, Mantsch JR. Glutamatergic plasticity in medial prefrontal cortex and ventral tegmental area following extended-access cocaine self-administration. Brain Res, 2011;1413:60-71.
Wang J, Yuan W, Li MD. Genes and Pathways Co-Associated With the Exposure to Multiple Drugs of Abuse, Including Alcohol, Amphetamine/Methamphetamine, Cocaine, Marijuana, Morphine, and/or Nicotine: A Review of Proteomics Analyses. Mol Nerobiol, 2011;44:269-86.
Sabol KE, Roach JT, Broom SL, Ferreira C, Preau MM. Long-term effects of a high-dose methamphetamine regimen on subsequent methamphetamine-induced dopamine release in vivo. Brain Res, 2001;892(1):122-9.
Nudmamud-Thanoi S, Iamjan SA, Kerdsan-Phusan W, Thanoi S. Pharmacogenetics of drug dependence: Polymorphisms of genes involved in glutamate neurotransmission. Neurosci Lett, 2019A;134128.
Nudmamud-Thanoi S, Veerasakul S, Thanoi S. Pharmacogenetics of drug dependence: Polymorphisms of genes involved in GABA neurotransmission. Neurosci Lett, 2019B;134463.
Chandramouli K, Qian PY. Proteomics: challenges, techniques and possibilities to overcome biological sample complexity. Hum Genomics Proteomics, 2009;2009:239204.
Li X, Wang H, Qiu P, Luo H. Proteomic Profiling of Proteins Associated With Methamphetamine-Induced Neurotoxicity in Different Regions of Rat Brain. Neurochem Int, 2008;52(1-2):256-64.
Faure JJ, Hattingh SM, Stein DJ, Daniels WM. Proteomic analysis reveals differentially expressed proteins in the rat frontal cortex after methamphetamine treatment. Metab Brain Dis, 2009;24(4):685-700.
Zhu R, Yang T, Kobeissy F, Mouhieddine TH, Raad M, Nokkari A, et al. The Effect of Chronic Methamphetamine Exposure on the Hippocampal and Olfactory Bulb Neuroproteomes of Rats. PLoS One, 2016;11(4):e0151034.
Scheiffele P. Cell-cell signaling during synapse formation in the CNS. Annu Rev Neurosci, 2003;26:485-508.
Segal DS, Kuczenski R, O'Neil ML, Melega WP, Cho AK. Escalating dose methamphetamine pretreatment alters the behavioral and neurochemical profiles associated with exposure to a high-dose methamphetamine binge. Neuropsychopharmacology, 2003;28(10):1730-40.
Veerasakul S, Thanoi S, Reynolds GP, Nudmamud-Thanoi S. Effect of Methamphetamine Exposure on Expression of Calcium Binding Proteins in Rat Frontal Cortex and Hippocampus. Neurotox Res, 2016;30(3):427-33.
Bosch PJ, Peng L, Kivell BM. Proteomics Analysis of Dorsal Striatum Reveals Changes in Synaptosomal Proteins following Methamphetamine Self-Administration in Rats. PLoS One, 2015;10(10):e0139829.
Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis, 1999;20:3551-67.
Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res, 2017;45(D1):D183-D189.
Szklarczyk D, Santos A, von Mering C, Jensen LJ, Bork P, Kuhn M. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res, 2016;44(D1):D380-D384.
Simões PF, Silva AP, Pereira FC, Marques E, Milhazes N, Borges F, et al. Methamphetamine changes NMDA and AMPA glutamate receptor subunit levels in the rat striatum and frontal cortex. Ann N Y Acad Sci, 2008;1139:232-41.
Kerdsan W, Thanoi S, Nudmamud-Thanoi S. Changes in glutamate/NMDA receptor subunit 1 expression in rat brain after acute and subacute exposure to methamphetamine. J Biomed Biotechnol, 2009;2009:329631.
Shigeri Y, Seal RP, Shimamoto K. Molecular pharmacology of glutamate transporters, EAATs and VGLUTs. Brain Res Brain Res Rev, 2004;45(3):250-65.
Iamjan SA, Thanoi S, Watiktinkorn P, Reynolds GP, Nudmamud-Thanoi S. Genetic variation of GRIA3 gene is associated with vulnerability to methamphetamine dependence and its associated psychosis. J Psychopharmacol, 2018;32(3):309-15.
Takeichi T, Hori O, Hattori T, Kiryu K, Zuka M, Kitamura O. Pre-administration of low-dose methamphetamine enhances movement and neural activity after high-dose methamphetamine administration in the striatum. Neurosci Lett, 2019;703:119-24.
Mao L, Wang JQ. Differentially altered mGluR1 and mGluR5 mRNA expression in rat caudate nucleus and nucleus accumbens in the development and expression of behavioral sensitization to repeated amphetamine administration. Synapse, 2001;41(3):230-40.
Bouarab C, Thompson B, Polter AM. VTA GABA Neurons at the Interface of Stress and Reward. Front Neural Circuits, 2019;13:78.
Impagnatiello F, Guidotti AR, Pesold C, Dwivedi Y, Caruncho H, Pisu MG, et al. A decrease of reelin expression as a putative vulnerability factor in schizophrenia. Proc Natl Acad Sci U S A, 1998;95(26):15718-23.
Guidotti A, Auta J, Davis JM, DiGiorgi Gerevini V, Dwivedi Y, Grayson DR, et al. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Arch Gen Psychiatry, 2000;57(11):1061-69.
Li H, Lu Q, Xiao E, Li Q, He Z, Mei X. Methamphetamine enhances the development of schizophrenia in first-degree relatives of patients with schizophrenia. Can J Psychiatry, 2014;59(2):107-13.
Veerasakul S, Watiktinkorn P, Thanoi S, Reynolds GP, Nudmamud-Thanoi S. Association of polymorphisms in GAD1 and GAD2 genes with methamphetamine dependence. Pharmacogenomics. 2017;18(1):17-22.
Kuriyama K, Hirouchi M, Kimura H. Neurochemical and molecular pharmacological aspects of the GABA(B) receptor. Neurochem Res, 2000;25(9-10):1233-39.
Mark KA, Soghomonian JJ, Yamamoto BK. High-dose methamphetamine acutely activates the striatonigral pathway to increase striatal glutamate and mediate long-term dopamine toxicity. J Neurosci, 2004;24(50):11449-56.
Ferrucci M, Limanaqi F, Ryskalin L, Biagioni F, Busceti CL, Fornai F. The Effects of Amphetamine and Methamphetamine on the Release of Norepinephrine, Dopamine and Acetylcholine From the Brainstem Reticular Formation. Front Neuroanat, 2019;13:48.
Picciotto MR, Higley MJ, Mineur YS. Acetylcholine as a neuromodulator: cholinergic signaling shapes nervous system function and behavior. Neuron, 2012;76(1):116-29.
Siegel JA, Craytor MJ, Raber J. Long-term effects of methamphetamine exposure on cognitive function and muscarinic acetylcholine receptor levels in mice. Behav Pharmacol, 2010;21(7):602-14.
Stoll K, Hart R, Lindsley CW, Thomsen M. Effects of muscarinic M1 and M4 acetylcholine receptor stimulation on extinction and reinstatement of cocaine seeking in male mice, independent of extinction learning. Psychopharmacology (Berl), 2018;235(3):815-27.
Sharma G, Vijayaraghavan S. Nicotinic Receptors: Role in Addiction and Other Disorders of the Brain. Subst Abuse, 2008;2008(1):81.
Brunzell DH, McIntosh JM. Alpha7 nicotinic acetylcholine receptors modulate motivation to self-administer nicotine: implications for smoking and schizophrenia. Neuropsychopharmacology, 2012;37(5):1134-43.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2021 Naresuan Phayao Journal

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
ผู้นิพนธ์ต้องรับผิดชอบข้อความในบทนิพนธ์ของตน มหาวิทยาลัยพะเยาไม่จำเป็นต้องเห็นด้วยกับบทความที่ตีพิมพ์เสมอไป ผู้สนใจสามารถคัดลอก และนำไปใช้ได้ แต่จะต้องขออนุมัติเจ้าของ และได้รับการอนุมัติเป็นลายลักษณ์อักษรก่อน พร้อมกับมีการอ้างอิงและกล่าวคำขอบคุณให้ถูกต้องด้วย
The authors are themselves responsible for their contents. Signed articles may not always reflect the opinion of University of Phayao. The articles can be reproduced and reprinted, provided that permission is given by the authors and acknowledgement must be given.







