Development of noninvasive prenatal testing of down’s syndrome by droplet digital PCR
Keywords:
Down’s syndrome, Prenatal diagnosis, NIPT, ddPCRAbstract
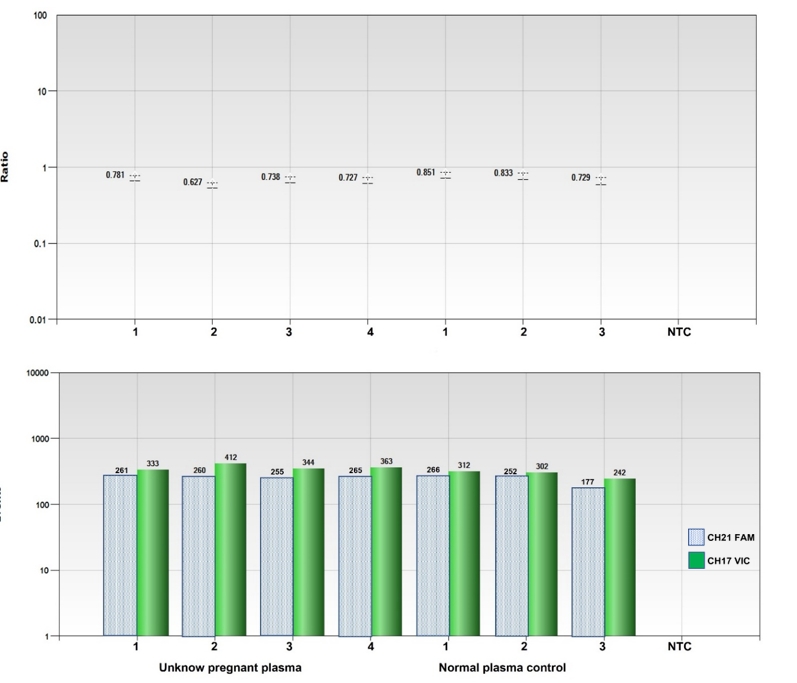
Down’s syndrome caused by aneuploidy of chromosome 21 (trisomy 21: T21) has high prevalence. Nowadays, advanced DNA technologies are used in noninvasive prenatal testing (NIPT) to screen for numerical chromosomal abnormalities in pregnancy at advanced maternal age. These technologies are expensive and complicated. The aim of this study is to develop the droplet digital PCR (ddPCR) technique for NIPT of T21. DdPCR condition was optimized by using DNA samples extracted from blood of children with T21, blood and plasma of non-pregnant normal women. Then, plasma cell-free DNA (cfDNA) from 30 pregnant women was tested using optimized ddPCR. Ratio of mutant allele/wild type allele was calculated by Bio-Rad QuantaSoft Analysis program. The ratios of blood and plasma of non-pregnant women were 1.043±0.073 and 0.710±0.019, respectively. The ratio of T21 patients was 1.486±0.107. The ratio of negative control plasma samples was 0.705±0.074. The result of cfDNA from plasma of 29 pregnant women was 0.742±0.062 which meant that there was a low probability to have T21 fetuses. The results of NIPT in 29 cfDNA samples were the same as those of cytogenetic methods. One cfDNA sample was excluded because of vanishing twin syndrome. Thus, this developed ddPCR-based NIPT was able to detect a low-risk group for T21; however, more pregnancy plasma samples are needed in order to find pregnant women with high risk of having T21 fetuses.
References
Pangkanon S, Layangool T, Niramis R, Keyurapan B, Intakorn P, Fuengfoo A, et al. Multidisciplinary approach in Down syndrome. Thai Pediatr J. 2008;15(2):227-31.
Asim A, Kumar A, Muthuswamy S, Jain S, Agarwal S. "Down syndrome: an insight of the disease". J Biomed Sci. 2015;22(1):41. doi: 10.1186/s12929-015-0138-y.
Mai CT, Isenburg JL, Canfield MA, Meyer RE, Correa A, Alverson CJ, Lupo PJ, Riehle-Colarusso T, Cho SJ, Aggarwal D, Kirby RS; National Birth Defects Prevention Network. National population-based estimates for major birth defects, 2010-2014. Birth Defects Res. 2019;111(18):1420-1435. doi: 10.1002/bdr2.1589.
Jaruratanasirikul S, Kor-Anantakul O, Chowvichian M, Limpitikul W, Dissaneevate P, Intharasangkanawin N, Sattapanyo A, Pathompanitrat S, Sriplung H. A population-based study of prevalence of Down syndrome in Southern Thailand. World J Pediatr. 2017;13(1):63-69. doi: 10.1007/s12519-016-0071-5.
Wongkrajang P, Jittikoon J, Sangroongruangsri S, Talungchit P, Ruangvutilert P, Panchalee T, Chaikledkaew U. Prenatal screening tests and prevalence of fetal aneuploidies in a tertiary hospital in Thailand. PLoS One. 2023;18(4):e0284829. doi: 10.1371/journal.pone.0284829.
Beazoglou T, Heffley D, Kyriopoulos J, Vintzileos A, Benn P. Economic evaluation of prenatal screening for Down syndrome in the U.S.A. Prenat Diagn 1998;18:1241-52.
ACOG Practice Bulletin. Clinical Management Guidelines for Obstetrician-Gynecologists. Prenatal diagnosis of fetal chromosomal abnormalities. Obstet Gynecol. 2001;97: suppl 1-12.
Pattanaphesaj J, Tonmukayakul U, Teerawattananon Y. Cost-benefit Analysis of Prenatal Screening and Diagnosis for Down Syndrome in Thailand. J Health Sci 2017;21(4):667-84.
Alfirevic Z, Neilson JP. Antenatal screening for Down's syndrome. BMJ. 2004;329(7470):811-2. doi: 10.1136/bmj.329.7470.811.
Benn PA. Advances in prenatal screening for Down syndrome: I. general principles and second trimester testing. Clin Chim Acta. 2002;323(1-2):1-16. doi: 10.1016/s0009-8981(02)00186-9.
Seeds JW. Diagnostic mid trimester amniocentesis: how safe?. Am J Obstet Gynecol 2004;191(2):607-15.
ACOG Practice Bulletin No. 77: screening for fetal chromosomal abnormalities. Obstet Gynecol 2007; 109:217-27.
Akolekar R, Beta J, Picciarelli G, Ogilvie C, D’Antonio F. Procedure-related risk of miscarriage following amniocentesis and chorionic villus sampling: a systematic review and meta-analysis. Ultrasound Obstet Gynecol. 2015;45:16–26.
Stomornjak-Vukadin M, Kurtovic-Basic I, Mehinovic L, Konjhodzic R. Combined use of cytogenetic and molecular methods in prenatal diagnostics of chromosomal abnormalities. Acta Inform Med. 2015;23(2):68-72. doi: 10.5455/aim.2015.23.68-72.
Gekas J, van den Berg DG, Durand A, Vallée M, Wildschut HI, Bujold E, et al. Rapid testing versus karyotyping in Down's syndrome screening: cost-effectiveness and detection of clinically significant chromosome abnormalities. Eur J Hum Genet. 2011;19(1):3-9. doi: 10.1038/ejhg.2010.138.
Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350(9076):485-7. doi: 10.1016/S0140-6736(97)02174-0.
Rafi I, Hill M, Hayward J, Chitty LS. Non-invasive prenatal testing: use of cell-free fetal DNA in Down syndrome screening. Br J Gen Pract. 2017;67(660):298-299. doi: 10.3399/bjgp17X691625.
ACOG American College of Obstetricians and Gynecologists Committee on Genetics. Committee Opinion No. 545: noninvasive prenatal testing for fetal aneuploidy. Obstet Gyn. 2012;120:1532-4
Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci U S A. 2008;105(42):16266-71. doi: 10.1073/pnas.0808319105.
Mersy E, Smits LJ, van Winden LA, de Die-Smulders CE; South-East Netherlands NIPT Consortium; Paulussen AD, Macville MV, Coumans AB, Frints SG. Noninvasive detection of fetal trisomy 21: systematic review and report of quality and outcomes of diagnostic accuracy studies performed between 1997 and 2012. Hum Reprod Update. 2013;19(4):318-29. doi: 10.1093/humupd/dmt001.
Chitty LS, Lo YM. Noninvasive Prenatal Screening for Genetic Diseases Using Massively Parallel Sequencing of Maternal Plasma DNA. Cold Spring Harb Perspect Med. 2015;5(9):a023085. doi: 10.1101/cshperspect.a023085.
Stokowski R, Wang E, White K, Batey A, Jacobsson B, Brar H, et al. Clinical performance of non-invasive prenatal testing (NIPT) using targeted cell-free DNA analysis in maternal plasma with microarrays or next generation sequencing (NGS) is consistent across multiple controlled clinical studies. Prenat Diagn. 2015;35(12):1243-6. doi: 10.1002/pd.4686.
Nykel A, Kaszkowiak M, Fendler W, Gach A. Chip-Based Digital PCR Approach Provides A Sensitive and Cost-Effective Single-Day Screening Tool for Common Fetal Aneuploidies-A Proof of Concept Study. Int J Mol Sci. 2019;20(21):5486. doi: 10.3390/ijms20215486.
Tan C, Chen X, Wang F, Wang D, Cao Z, Zhu X, et al. A multiplex droplet digital PCR assay for non-invasive prenatal testing of fetal aneuploidies. Analyst. 2019;144:2239–2247.
Dai P, Yang Y, Zhao G, Gu Z, Ren H, Hu S, et al. A dPCR-NIPT assay for detections of trisomies 21, 18 and 13 in a single-tube reaction-could it replace serum biochemical tests as a primary maternal plasma screening tool? J Transl Med. 2022;20(1):269. doi: 10.1186/s12967-022-03455-y.
Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Analysis of the size distributions of fetal and maternal cell-free DNA by paired-end sequencing. Clin Chem. 2010;56(8):1279-86. doi: 10. 1373/clinchem.2010.144188.
Yu SC, Chan KC, Zheng YW, Jiang P, Liao GJ, Sun H, et al. Size-based molecular diagnostics using plasma DNA for noninvasive prenatal testing. Proc Natl Acad Sci U S A. 2014;111(23):8583-8. doi: 10.1073/pnas.1406103111.
Hu P, Liang D, Chen Y, Lin Y, Qiao F, Li H, Wang T, Peng C, Luo D, Liu H, Xu Z. An enrichment method to increase cell-free fetal DNA fraction and significantly reduce false negatives and test failures for non-invasive prenatal screening: a feasibility study. J Transl Med. 2019;17(1):124. doi: 10.1186/s12967-019-1871-x.
Chiu RWK, Lo YMD. Cell-free fetal DNA coming in all sizes and shapes. Prenat Diagn. 2021;41(10):1193-1201. doi: 10.1002/pd.5952. Epub 2021 May 7.
Qi T, Pan M, Shi H, Wang L, Bai Y, Ge Q. Cell-Free DNA Fragmentomics: The Novel Promising Biomarker. Int J Mol Sci. 2023;24(2):1503. doi: 10.3390/ijms24021503.
Smith M, Lewis KM, Holmes A, Visootsak J. A Case of False Negative NIPT for Down Syndrome-Lessons Learned. Case Rep Genet. 2014;2014:823504. doi: 10.1155/2014/823504.
Lebo RV, Novak RW, Wolfe K, Michelson M, Robinson H, Mancuso MS. Discordant circulating fetal DNA and subsequent cytogenetics reveal false negative, placental mosaic, and fetal mosaic cfDNA genotypes. J Transl Med. 2015;13:260. doi: 10.1186/s12967-015-0569-y.
Van Opstal D, Srebniak MI, Polak J, de Vries F, Govaerts LC, Joosten M, Go AT, et al. False Negative NIPT Results: Risk Figures for Chromosomes 13, 18 and 21 Based on Chorionic Villi Results in 5967 Cases and Literature Review. PLoS One. 2016;11(1):e0146794. doi: 10.1371/journal.pone.0146794.
Xu HH, Dai MZ, Wang K, Zhang Y, Pan FY, Shi WW. A rare Down syndrome foetus with de novo 21q;21q rearrangements causing false negative results in non-invasive prenatal testing: a case report. BMC Med Genomics. 2020;13(1):96. doi: 10.1186/s12920-020-00751-8.
Liehr T. False-positives and false-negatives in non-invasive prenatal testing (NIPT): what can we learn from a meta-analyses on > 750,000 tests? Mol Cytogenet. 2022;15(1):36. doi: 10.1186/s13039-022-00612-2.
Kleinfinger P, Luscan A, Descourvieres L, Buzas D, Boughalem A, Serero S, et al. Noninvasive Prenatal Screening for Trisomy 21 in Patients with a Vanishing Twin. Genes (Basel). 2022;13(11):2027. doi: 10.3390/genes13112027.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2023 University of Phayao

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
ผู้นิพนธ์ต้องรับผิดชอบข้อความในบทนิพนธ์ของตน มหาวิทยาลัยพะเยาไม่จำเป็นต้องเห็นด้วยกับบทความที่ตีพิมพ์เสมอไป ผู้สนใจสามารถคัดลอก และนำไปใช้ได้ แต่จะต้องขออนุมัติเจ้าของ และได้รับการอนุมัติเป็นลายลักษณ์อักษรก่อน พร้อมกับมีการอ้างอิงและกล่าวคำขอบคุณให้ถูกต้องด้วย
The authors are themselves responsible for their contents. Signed articles may not always reflect the opinion of University of Phayao. The articles can be reproduced and reprinted, provided that permission is given by the authors and acknowledgement must be given.







