The Quality Evaluation of Clostridium difficile Toxin A/B Test Kit by Lateral Flow Immunochromatography Method
Main Article Content
Abstract
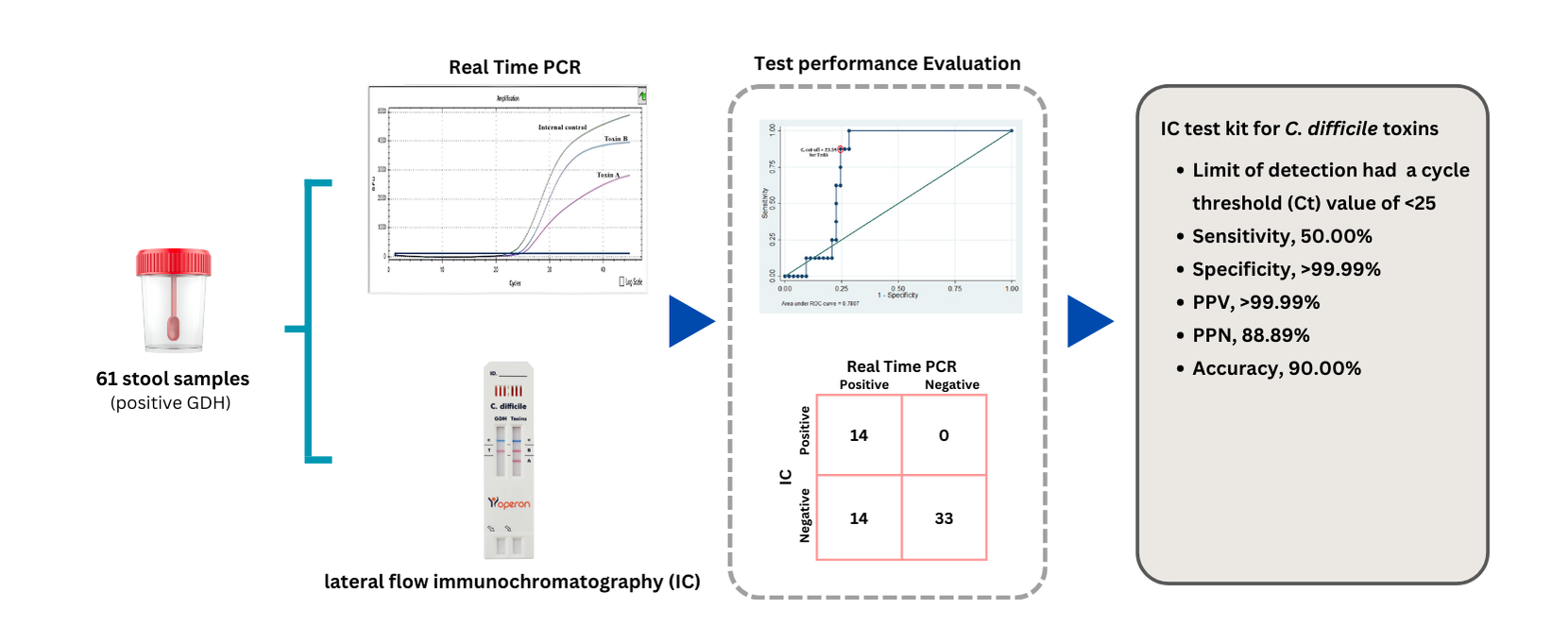
Clostridium (or Clostridiodes) difficile produces both toxins A (TcdA) and B (TcdB), which cause severe diarrhea in patients with C. difficile infection (CDI) and can lead to death. Diagnosis of glutamate dehydrogenase enzymes (GDH) and TcdA/B detection by lateral flow Immunochromatography (IC) test kits requires quality evaluation prior to clinical use. This study aimed to evaluate the performance of an IC test kit by comparing its results with those of real-time polymerase chain reaction (real-time PCR). Methods: A total of 61 fecal post-analysis samples from patients who attended Thammasat University Hospital, Pathum Thani, Thailand, were tested for GDH, TcdA/B using the IC test kit, with results confirmed by real-time PCR. Result: Real-time PCR confirmed that all samples were positive for C. difficile (100.00%). The distribution of toxigenic strains was as follows: TcdA+/TcdB+ (37.70%), TcdA-/TcdB+ (8.20%), and non-toxigenic strains (TcdA-/TcdB-) (54.10%). The IC test kit showed the following performance metrics for TcdA/B detection in CDI cases: sensitivity 50.00%, specificity >99.99%, positive predictive value (PPV) >99.99%, negative predictive value (NPV) 88.89%, and overall accuracy 90.00%. The cycle threshold (Ct) values for TcdA and TcdB were 23.59 and 24.43, respectively. Conclusion: The IC test kit for GDH and TcdA/B detection demonstrated acceptable quality for routine laboratory screening of C. difficile infection. However, confirmation using real-time PCR is recommended for accurate diagnosis.
Article Details
References
Bacci, S., Mølbak, K., Kjeldsen, M. K. and Olsen,K.E.,2011,Binary toxinand death after Clostridium difficile infection, Emerg. Infect. Dis. 17(6): 976–982.
Berry, C. E., Davies, K. A., Owens, D. W. and Wilcox, M. H., 2017, Is there a relationship between the presenceof the binary toxingenes in Clostridium difficile strains and the severity of C. difficile infection (CDI)?, Eur. J. Clin. Microbiol. Infect. Dis. 36(12): 2405–2415.
Imwattana, K., Wangroongsarb, P. and Riley, T. V., 2019, High prevalence and diversity of tcdA-negative and tcdB-positive, and non-toxigenic, Clostridium difficile in Thailand, Anaerobe. 57: 4–10.
Imwattana, K., Knight, D. R., Kullin, B., Collins, D. A. and Putsathit, P., 2019, Clostridium difficile ribotype 017 - characterization, evolution and epidemiology of the dominant strain in Asia, Emerg. Microbes. infect. 8(1): 796–807.
Sholeh, M., Kouhsari, E., Talebi, M., Hallajzadeh, M. and Godarzi, F., 2021, Toxin gene profiles and antimicrobial resistance of Clostridioides difficile infection: a single tertiary care center study in Iran, Iran. J. Microbiol. 13(6): 793–800.
Cohen, S. H., Tang, Y. J., Hansen, B. and Silva, J., Jr., 1998, Isolation of a toxin B-deficient mutant strain of Clostridium difficile in a case of recurrent C. difficileassociated diarrhea, Clin. Infect. Dis.26(2): 410–412.
Drudy, D., Harnedy, N., Fanning, S., O'Mahony,R.and Kyne,L.,2007, Isolation and characterizationof toxin A-negative, toxin B-positive Clostridium difficile in Dublin, Ireland, Clin. Microbiol. Infect. 13(3): 298–304.
Humphries,R. M., Uslan, D. Z. and Rubin, Z., 2013, Performance of Clostridium difficile toxinenzyme immunoassay and nucleic acid amplificationtests stratified by patient disease severity, J. Clin. Microbiol. 51(3): 869–873.
Operon Immuno and Molecular Diagnostics, 2025, Simple GDH-Toxins, Available Source :https://www.operondx.com/wp-content/uploads/pdf/instructions/0905144_GDH-Toxins_web.pdf, February 21, 2025.
Certest Biotec SL, Viasure Clostridium Difficile Real-Time PCR Detection Kit, Available Source :https://www.certest.es/wp-content/uploads/2015/12/VIASUREClostridium-difficile-EN.pdf, February 21,2024.
Certest Biotec SL, Viasure Clostridium Difficile Toxin A+B Real-Time PCR Detection Kit, Available Source: https://www.certest.es/wp-content/uploads/2015/12/VIASURE_ClostridiumdifficiletoxAB_EN.pdf,February21,2024.
Medcalc Software Ltd, Diagnostic Test Evaluation Calculator, Available Source: https://www.medcalc.org/calc/diagnostic_test.php (version22.021), May 9, 2024.
Nahm, F. S., 2022, Receiver operating characteristic curve: overview and practical use for clinicians, Korean J. Anesthesiol. 75(1): 25–36.
Polage, C. R., Gyorke, C. E., Kennedy, M. A., Leslie, J. L. and Chin, D. L., 2015, Overdiagnosis of Clostridium difficile infectioninthe molecular testera, JAMA. Intern. Med. 175(11): 1792–1801.
Putsathit,P., Kiratisin,P., Ngamwongsatit, P. and Riley, T. V., 2015, Clostridium difficile infection in Thailand, Intern. J. Antimicrob. Agents. 45(1): 1–7.
Wongwanich,S., Pongpech,P., Dhiraputra, C., Huttayananont,S. and Sawanpanyalert, P., 2001, Characteristics of Clostridium difficile strains isolated from asymptomatic individuals and from diarrheal patients, Clin. Microbiol. Infect.7(8): 438–441.
Putsathit, P., Maneerattanaporn, M., Piewngam,P., Kiratisin,P. and Riley,T.V., 2016, Prevalence and molecular epidemiology of Clostridium difficile infection in Thailand, New. Microbes. New. Infect. 15: 27–32.
Wongkuna, S., Janvilisri, T., Phanchana, M., Harnvoravongchai, P. and Aroonnual, A., 2021, Temporal variations in patterns of Clostridioides difficile strain diversity and antibiotic resistance in Thailand, Antibiotics. 10(6): 1-15.
Wongwanich, S., Ramsiri, S., Vanasin, B., Khowsaphit, P. and Tantipatayangkul, P., 1990, Clostridium difficile associated disease in Thailand, Southeast. Asian. J. Trop. Med. Pub. Health. 21(3): 367–372.
Issarachaikull, R., Khantipong, M., Sawatpanich, A. and Suankratay, C., 2015, Prospective evaluation of a novel two-step protocol for screeningof Clostridium difficile infection in hospitalized adult patients, Southeast. Asian. J. Trop. Med. Pub. Health. 46(6): 1037–1048.
Chapin,K. C., Dickenson,R. A., Wu,F.and Andrea, S. B., 2011, Comparison of five assays for detection of Clostridium difficile toxin, J. Mol. Diagn. 13(4): 395–400.
Jaqueti, A. J., Molina, E. L. M., García-Arata, I., García-Martínez, J. and Cano De Torres, I., 2021, Significance of a polymerase chain reaction method inthe detection of Clostridioides difficile, Rev. Esp. Quimioter. 34(2): 141–144.
Fedorko, D.P., Engler, H. D., O'Shaughnessy, E. M., Williams, E. C. and Reichelderfer, C. J., 1999, Evaluationof tworapid assays for detectionof Clostridium difficiletoxin A in stool specimens, J. Clin. Microbiol. 37(9): 3044–3047.
René, P., Frenette, C. P., Schiller, I., Dendukuri, N. and Brassard, P., 2012, Comparison of eight commercial enzyme immunoassays for the detection of Clostridium difficile from stool samples and effect of strain type, Diagn. Microb. Infect. Dis. 73: 94–96.
Eastwood, K., Else, P., Charlett, A. and Wilcox, M., 2009, Comparison of nine commercially available Clostridium difficile toxin detection assays, a real-time PCR assay for C. difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin testing and cytotoxigenic culture methods, J. Clin. Microb. 47(10): 3211–3217.
Burnham, C. A. and Carroll, K. C., 2013, Diagnosisof Clostridium difficileinfection: anongoing conundrum for clinicians and for clinical laboratories, Clin. Microbiol. Rev. 26(3): 604–630.
Alcalá, H. L., Reigadas, R. E. and Bouza, S. E., 2017, Clostridium difficileinfection, Med. Clin. 148(10): 456–463.
Lee, S., Nanda, N., Yamaguchi, K., Lee, Y. and She, R. C., 2022, Clostridioides difficile Toxin B pcr cycle threshold as a predictor of toxin testing in stool specimens from hospitalized adults, Antibiotics. 11(5): 1-9.