Analysis of the D-loop, Phe-tRNA, 12S-rRNA (bar2) mitochondrial gene in black-bone chickens compared to other chickens from gene banks
Main Article Content
Abstract
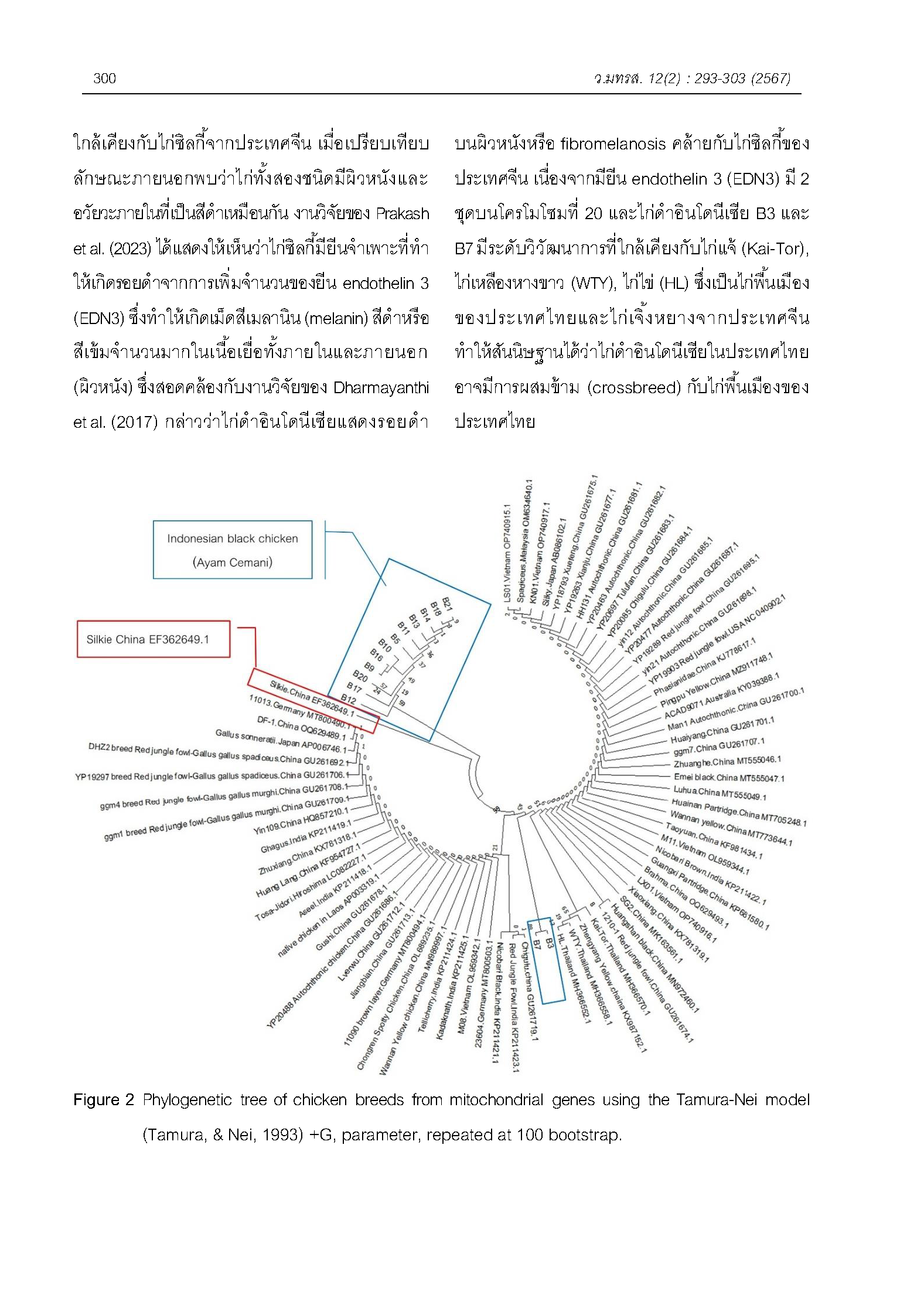
Black chickens are an economically important animal species that are extensively grown, having high nutritional value and therapeutic characteristics, which work as an antioxidant and play a role in immunological function. They are popular among health-conscious individuals. Black-bone chicken is a variety of chicken breeds raised in Thailand, while black chickens are expensive compared to other chicken breeds, especially Indonesian black chickens which are black throughout the body even the feathers, bones and meat. So, there is a demand for pure breed Indonesian black chickens. However, there is a lack of information on the genetic diversity of black-bone chickens raised in Thailand. This study aimed to find the genetic diversity of the D-loop, Phe-tRNA and 12S-rRNA (bar2) genes located in the mitochondria. The 14 Indonesian black chickens grown in Thailand were compared to 70 samples of chicken data from the GenBank database to identify genetic markers that differ in different chicken breeds. The analysis was performed using PCR and then the PCR product was analyzed to determine the base sequence. The haplotypes were determined using DnaSP6 and the phylogenetic tree was constructed using MEGA 11. The results found that most Indonesian black chickens had nucleotide sequences of their target genes that were uniquely different from those of other chickens (outgroup). On the other hand, they had nucleotide sequences that were closest to the Silkie chickens, based on the results shown in the phylogenetic tree. The results of the haplotype found that Indonesian black chickens were divided into 7 haplotypes, indicating the high genetic diversity of Indonesian black chickens in Thailand. It also was found that the sequence at position 812 bp had a specific sequence in haplotypes 1-7 of Indonesian black chickens with T bases, while haplotypes 8-14 of chickens from the GenBank database had all C bases. In future study, the nucleotide sequence of Indonesian chickens at the 812 bp location, specifically the SNPs, can be utilized as a molecular marker to identify and select black chickens.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Published manuscript are the rights of their original owners and RMUTSB Academic Journal. The manuscript content belongs to the authors' idea, it is not the opinion of the journal's committee and not the responsibility of Rajamangala University of Technology Suvarnabhumi
References
Arif, I. A., Khan, H. A., Bahkali, A. H., Al Homaidan, A. A., Al Farhan, A. H., Al Sadoon, M., & Shobrak, M. (2011). DNA marker technology for wildlife conservation. Saudi Journal of Biological Sciences, 18(3), 219-225.
Bao, H., Zhao, C., Li, J., & Wu, C. (2008). Sequencing and alignment of mitochondrial genomes of Tibetan chicken and two lowland chicken breeds. Science in China Series C: Life Sciences, 51, 47-51.
Cui, H., Ibtisham, F., Xu, C., Huang, H., & Su, Y. (2017). DNA barcoding of Chinese native chicken breeds through COI gene. The Thai Journal of Veterinary Medicine, 47(1), 123-129.
Daetwyler, H. D., Villanueva B., Bijma P., & Woolliams J. A. (2007). Inbreeding in genome-wide selection. Journal of Animal Breeding and Genetics, 124(6), 369-376.
Desjardins, P., & Morais, R. (1990). Sequence and gene organization of the chicken mitochondrial genome:
a novel gene order in higher vertebrates. Journal of Molecular Biology, 212(4), 599-634.
Dharmayanthi, A. B., Terai, Y., Sulandari, S., Zein, M. S. A., Akiyama, T., & Satta, Y. (2017). The origin and evolution of fibromelanosis in domesticated chickens: genomic comparison of Indonesian cemani and Chinese silkie breeds. PLOS ONE, 12(4), e0173147.
Di Lorenzo, P., Ceccobelli, S., Panella, F., Attard, G., & Lasagna, E. (2015). The role of mitochondrial DNA to determine the origin of domestic chicken. World's Poultry Science Journal, 71(2), 311-318.
Dorshorst, B., Okimoto, R., & Ashwell, C. (2010). Genomic regions associated with dermal hyperpigmentation, polydactyly and other morphological traits in the Silkie chicken. Journal of Heredity, 101(3), 339-350.
Groeneveld, L. F., Lenstra, J. A., Eding, H., Toro, M. A., Scherf, B., Pilling, D., Negrini, R., Finlay, E. K., Jianlin, H., Groeneveld, E., Weigend, S., & The GLOBALDIV Consortium. (2010). Genetic diversity in farm animals--a review. Animal Genetics, 41(s1), 6-31.
Habsari, I. K., Nugroho, B. A., & Azizah, S. (2018). Supply chain analysis of cemani chickens in Temanggung, Central Java, Indonesia. IOSR Journal of Economics and Finance, 9(4), 44-49.
Kostaman, T., Sopiyana, S., Isbandi, & Pasaribu, T. (2021). Ex-situ exploration of cemani chicken in Balai Penelitian Ternak (Balitnak), Bogor-West Java. BIO Web of Conferences, 33, 01001.
Lan, D., Hu, Y., Zhu, Q., & Liu, Y. (2017). Mitochondrial DNA study in domestic chicken. DNA Mapping, Sequencing, and Analysis, 28(1), 25-29.
Likittrakulwong, W., Poolprasert, P., & Roytrakul, S. (2018). Morphological trait, molecular genetic evidence
and proteomic determination of different chickens (Gallus gallus) breeds. Journal of Applied Biology & Biotechnology, 7(1), 65-70.
Lukanov, H., & Genchev, A. (2013). Fibromelanosis in domestic chickens. Journal of Agricultural Science and Technology, 5(3), 239-246.
Meerasen, S., Duangjit, S., & Yasothonsrikul, S. (2009). Analysis of the mutation rate of the mitochondrial DNA region. hypervariable region in the lower northern region of Thailand (research report). Phitsanulok: Naresuan University. (in Thai)
Miao, Y. W., Peng, M. S., Wu, G. S., Ouyang, Y. N., Yang, Z. Y., Yu, N., Liang, J. P., Pianchou, G., Beja-Pereira, A., Mitra, B., Palanichamy, M. G., Baig, M., Chaudhuri, T. K., Shen, Y. Y., Kong, Q. P., Murphy, R. W., Yao, Y. G., & Zhang, Y. P. (2013). Chicken domestication: an updated perspective based on mitochondrial genomes. Heredity, 110, 277-282.
Muladno, M. (2008). Local chicken genetic resources and production systems in Indonesia. Retrieved 21 October 2023, from http://www.fao.org/docrep/013/al695e/al695e00.pdf.
Muroya, S., Tanabe, R. I., Nakajima, I., & Chikuni, K. (2000). Molecular characteristics and site specific distribution of the pigment of the Silky fowl. Journal of Veterinary Medical Science, 62(4), 391-395.
Prakash, A., Singh, Y., Chatli, M. K., Sharma, A., Acharya, P., & Singh M. (2023). Review of the black meat chicken breeds: Kadaknath, Silkie, and Ayam Cemani. World's Poultry Science Journal, 79(4), 879-891.
Robertson, D., Shore, S., & Miller, D. M. (1997). Manipulation and expression of recombinant DNA; a laboratory manual. Raleigh (USA): Academic Press.
Rozas, J., Ferrer-Mata, A., Sanchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., & Sanchez-Gracia, A. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution, 34(12), 3299-3302.
Siriwadee, P., Wirot L., Thanapol P., & Wirawan N. (2023). Genetic diversity among five native Thai chickens and Khiew-Phalee chickens in lower-northern Thailand using mitochomdrial DNA barcodes. Biodiversitas, 24(4), 1962-1970.
Shi, H., Fu, J., He, Y., Li, Z., Kang, J., Hu, C., Zi, X., Liu, Y., Zhao, J., Dou, T., Jia, J., Duan, Y., Wang, K., & Ge, C. (2022). Hyperpigmentation inhibits early skeletal muscle development in Tengchong snow chicken breed. Genes, 13(12), 2253.
Tamura, K., & Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution, 10(3), 512-526.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., & Kumar, S. (2011). MEGA5: molecular evolutionary genetic analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28(10), 2731-2739.
Wilkinson, S., Wiener, P., Teverson, D., Haley, C. S., & Hocking, P. M. (2012). Characterization of the genetic diversity, structure and admixture of British chicken breeds. Animal Genetics, 43(5), 552-563.
Yu, S., Wang, G., Liao, J., & Tang, M. (2018). Transcriptome profile analysis identifies candidate genes for the melanin pigmentation of breast muscle in Muchuan black-boned chicken. Poultry Science, 97(10), 3446-3455.
Wolc, A., Stricker C., Arango J., Settar P., Fulton J. E., O’Sullivan N. P., Preisinger R., Habier D., Fernando R., Garrick D. J., Lamont S. J., & Dekkers J. C. M. (2011). Breeding value prediction for production traits in layer chickens using pedigree or genomic relationships in a reduced animal model. Genetics Selection Evoluation, 43, 5.